Gene
KWMTBOMO01690
Pre Gene Modal
BGIBMGA006034
Annotation
calcium_and_integrin_binding_protein_CIB_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.8
Sequence
CDS
ATGGGTCAAGGAAAAAGCCAATTTACAGAAGAAGAACTACAAGACTATGAGGATCTCACGTACTTTACGAAGAAAGAAGTATTGTATGCTCACCAAAAATTCAAAGCTCTGGCACCTGAGAAAGTTGGGCACAATAAAAATGCTAAATTACCTATGGCCAAAGTGCTCACATATCCAGAACTTAAAGTGAATCCATTCAAAGATAGGATATGTAAGGTGTTCAGTTCCAGTAATGATGGAGATTGCACCTTTGAAGACTTCTTGGACATGATGTCAGTTTTCAGTGAGATGGCTCCTAAAGCAGTAAAAGCTGAACATGCATTTAGGATATTTGATTTTGATGGTGATGACATGATAGGCGTATCAGATTTACGTGAAGTAATACAGCGTCTGTGCGGACCGGAGTTAAAACTATCAGACTCAGAAATACAACAACTGGTGCAGAATGTTTTGGAAGAAGCAGATCTTGATGATGATGGAGCTCTCTCCTTTGCTGAGTTTGAACATATCATAGATAAGAGTTCTGATTTCTGCCATGCATTCCGAATCAGGCTTTAA
Protein
MGQGKSQFTEEELQDYEDLTYFTKKEVLYAHQKFKALAPEKVGHNKNAKLPMAKVLTYPELKVNPFKDRICKVFSSSNDGDCTFEDFLDMMSVFSEMAPKAVKAEHAFRIFDFDGDDMIGVSDLREVIQRLCGPELKLSDSEIQQLVQNVLEEADLDDDGALSFAEFEHIIDKSSDFCHAFRIRL
Summary
Uniprot
Q2F616
A0A2W1BJA8
A0A2A4KAH6
A0A1S6M472
I4DNU1
S4PM05
+ More
H9J942 A0A0L7LH95 A0A194R5Z1 A0A151WYP7 A0A158P2Z9 A0A067REN5 E2AUN1 A0A1B6FR13 A0A0T6B2I7 A0A1B6KFG0 A0A1Y1KNI6 A0A139WI15 V5H0Y6 A0A3L8DQV1 A0A212EJ40 A0A232F8E0 K7J3T7 E0VJM0 A0A1B6CPG4 A0A0L7K2G8 A0A0K8TS38 A0A195CEE0 A0A195DPF2 F4X5F4 A0A195BNE3 A0A195EZ36 A0A0J7L569 A0A1B0GMU7 A0A1B0GKG0 A0A0C9RSC9 A0A2H1WN62 A0A1L8DW92 A0A2J7QVV7 Q17IW0 A0A182LUX7 A0A182RRL5 A0A182WHG1 A0A023EI62 N6TET7 A0A026W121 A0A2M4C0S8 A0A182VHW9 A0A182TKX6 A0A182X145 A0A182L093 Q7Q4L8 A0A182IBY3 J3JZ87 A0A2A3EML9 A0A2M3ZJD8 A0A2M4CTB6 A0A0L7QK37 A0A1W4WK49 A0A182QZS5 A0A182K5H8 A0A182P661 A0A088ADJ0 A0A1Q3FGD1 B0W8S6 A0A2M4AUK2 U5EZE3 A0A0V0G8A0 R4WNU4 A0A023FCK3 T1HPB1 A0A336M077 A0A1S3CTK2 A0A0A9XL40 E2BRZ0 A0A0P4VSZ7 A0A1W4WJ65 A0A182NUU2 A0A0A1WJF2 A0A0K8W1T1 A0A034W8E1 A0A182Y5I2 T1DQY5 W8BS87 A0A1L8DLT9 A0A1B6HXZ2 A0A1J1HRE3 A0A0N0BEQ2 A0A182GYS5 A0A182F2B9 A0A1B6KWN8 J9K310 C4WVJ8 A0A0K8R8E5 A0A3M6U3X9 A0A2P2IB89 R7T7Q4 A7RM01 A0A210PED7 A0A1E1XQU4
H9J942 A0A0L7LH95 A0A194R5Z1 A0A151WYP7 A0A158P2Z9 A0A067REN5 E2AUN1 A0A1B6FR13 A0A0T6B2I7 A0A1B6KFG0 A0A1Y1KNI6 A0A139WI15 V5H0Y6 A0A3L8DQV1 A0A212EJ40 A0A232F8E0 K7J3T7 E0VJM0 A0A1B6CPG4 A0A0L7K2G8 A0A0K8TS38 A0A195CEE0 A0A195DPF2 F4X5F4 A0A195BNE3 A0A195EZ36 A0A0J7L569 A0A1B0GMU7 A0A1B0GKG0 A0A0C9RSC9 A0A2H1WN62 A0A1L8DW92 A0A2J7QVV7 Q17IW0 A0A182LUX7 A0A182RRL5 A0A182WHG1 A0A023EI62 N6TET7 A0A026W121 A0A2M4C0S8 A0A182VHW9 A0A182TKX6 A0A182X145 A0A182L093 Q7Q4L8 A0A182IBY3 J3JZ87 A0A2A3EML9 A0A2M3ZJD8 A0A2M4CTB6 A0A0L7QK37 A0A1W4WK49 A0A182QZS5 A0A182K5H8 A0A182P661 A0A088ADJ0 A0A1Q3FGD1 B0W8S6 A0A2M4AUK2 U5EZE3 A0A0V0G8A0 R4WNU4 A0A023FCK3 T1HPB1 A0A336M077 A0A1S3CTK2 A0A0A9XL40 E2BRZ0 A0A0P4VSZ7 A0A1W4WJ65 A0A182NUU2 A0A0A1WJF2 A0A0K8W1T1 A0A034W8E1 A0A182Y5I2 T1DQY5 W8BS87 A0A1L8DLT9 A0A1B6HXZ2 A0A1J1HRE3 A0A0N0BEQ2 A0A182GYS5 A0A182F2B9 A0A1B6KWN8 J9K310 C4WVJ8 A0A0K8R8E5 A0A3M6U3X9 A0A2P2IB89 R7T7Q4 A7RM01 A0A210PED7 A0A1E1XQU4
Pubmed
28756777
22651552
26354079
23622113
19121390
26227816
+ More
21347285 24845553 20798317 28004739 18362917 19820115 30249741 22118469 28648823 20075255 20566863 26369729 21719571 17510324 24945155 23537049 24508170 20966253 12364791 14747013 17210077 22516182 23691247 25474469 25401762 26823975 27129103 25830018 25348373 25244985 24495485 26483478 30382153 23254933 17615350 28812685 29209593
21347285 24845553 20798317 28004739 18362917 19820115 30249741 22118469 28648823 20075255 20566863 26369729 21719571 17510324 24945155 23537049 24508170 20966253 12364791 14747013 17210077 22516182 23691247 25474469 25401762 26823975 27129103 25830018 25348373 25244985 24495485 26483478 30382153 23254933 17615350 28812685 29209593
EMBL
DQ311256
ABD36201.1
KZ150052
PZC74391.1
NWSH01000011
PCG80914.1
+ More
KY460554 AQU15098.1 AK403139 KQ459584 BAM19581.1 KPI98517.1 GAIX01003785 JAA88775.1 BABH01004466 JTDY01001197 KOB74571.1 KQ460644 KPJ13238.1 KQ982649 KYQ52999.1 ADTU01007655 KK852514 KDR22217.1 GL442829 EFN62878.1 GECZ01017190 JAS52579.1 LJIG01016213 KRT81277.1 GEBQ01029818 GEBQ01010974 JAT10159.1 JAT29003.1 GEZM01080833 JAV62031.1 KQ971343 KYB27465.1 GALX01001973 JAB66493.1 QOIP01000005 RLU22563.1 AGBW02014536 OWR41495.1 NNAY01000665 OXU27101.1 DS235226 EEB13576.1 GEDC01022027 GEDC01011251 JAS15271.1 JAS26047.1 JTDY01017560 KOB51844.1 GDAI01000637 JAI16966.1 KQ977935 KYM98596.1 KQ980713 KYN14364.1 GL888716 EGI58310.1 KQ976432 KYM87550.1 KQ981905 KYN33478.1 LBMM01000704 KMQ97698.1 AJVK01004164 AJWK01028824 GBYB01010301 GBYB01010302 JAG80068.1 JAG80069.1 ODYU01009823 SOQ54505.1 GFDF01003395 JAV10689.1 NEVH01009771 PNF32702.1 CH477236 EAT46633.1 AXCM01003806 GAPW01004816 JAC08782.1 APGK01040757 KB740984 KB631904 ENN76278.1 ERL87104.1 KK107526 EZA49281.1 GGFJ01009752 MBW58893.1 AAAB01008964 EAA12727.2 APCN01000760 BT128568 AEE63525.1 KZ288206 PBC33053.1 GGFM01007844 MBW28595.1 GGFL01004418 MBW68596.1 KQ415041 KOC58881.1 AXCN02000745 GFDL01008436 JAV26609.1 DS231860 EDS39305.1 GGFK01011156 MBW44477.1 GANO01000952 JAB58919.1 GECL01001957 JAP04167.1 AK417336 BAN20551.1 GBBI01000194 JAC18518.1 ACPB03014848 UFQT01000240 SSX22243.1 GBHO01028766 GBHO01028763 GBHO01022962 GBHO01022960 GBRD01012148 GBRD01012146 GBRD01012145 GBRD01001216 GDHC01004344 JAG14838.1 JAG14841.1 JAG20642.1 JAG20644.1 JAG53676.1 JAQ14285.1 GL450107 EFN81506.1 GDKW01000970 JAI55625.1 GBXI01015315 JAC98976.1 GDHF01007245 JAI45069.1 GAKP01008909 JAC50043.1 GAMD01002194 JAA99396.1 GAMC01002335 JAC04221.1 GFDF01006774 JAV07310.1 GECU01028182 GECU01012705 JAS79524.1 JAS95001.1 CVRI01000010 CRK89046.1 KQ435824 KOX72231.1 JXUM01098082 KQ564393 KXJ72343.1 GEBQ01024132 JAT15845.1 ABLF02028424 AK341548 BAH71918.1 GADI01006371 JAA67437.1 RCHS01002300 RMX48296.1 IACF01005695 LAB71278.1 AMQN01014763 AMQN01014764 KB311248 ELT89640.1 DS469519 EDO47515.1 NEDP02076749 OWF34844.1 GFAA01001759 JAU01676.1
KY460554 AQU15098.1 AK403139 KQ459584 BAM19581.1 KPI98517.1 GAIX01003785 JAA88775.1 BABH01004466 JTDY01001197 KOB74571.1 KQ460644 KPJ13238.1 KQ982649 KYQ52999.1 ADTU01007655 KK852514 KDR22217.1 GL442829 EFN62878.1 GECZ01017190 JAS52579.1 LJIG01016213 KRT81277.1 GEBQ01029818 GEBQ01010974 JAT10159.1 JAT29003.1 GEZM01080833 JAV62031.1 KQ971343 KYB27465.1 GALX01001973 JAB66493.1 QOIP01000005 RLU22563.1 AGBW02014536 OWR41495.1 NNAY01000665 OXU27101.1 DS235226 EEB13576.1 GEDC01022027 GEDC01011251 JAS15271.1 JAS26047.1 JTDY01017560 KOB51844.1 GDAI01000637 JAI16966.1 KQ977935 KYM98596.1 KQ980713 KYN14364.1 GL888716 EGI58310.1 KQ976432 KYM87550.1 KQ981905 KYN33478.1 LBMM01000704 KMQ97698.1 AJVK01004164 AJWK01028824 GBYB01010301 GBYB01010302 JAG80068.1 JAG80069.1 ODYU01009823 SOQ54505.1 GFDF01003395 JAV10689.1 NEVH01009771 PNF32702.1 CH477236 EAT46633.1 AXCM01003806 GAPW01004816 JAC08782.1 APGK01040757 KB740984 KB631904 ENN76278.1 ERL87104.1 KK107526 EZA49281.1 GGFJ01009752 MBW58893.1 AAAB01008964 EAA12727.2 APCN01000760 BT128568 AEE63525.1 KZ288206 PBC33053.1 GGFM01007844 MBW28595.1 GGFL01004418 MBW68596.1 KQ415041 KOC58881.1 AXCN02000745 GFDL01008436 JAV26609.1 DS231860 EDS39305.1 GGFK01011156 MBW44477.1 GANO01000952 JAB58919.1 GECL01001957 JAP04167.1 AK417336 BAN20551.1 GBBI01000194 JAC18518.1 ACPB03014848 UFQT01000240 SSX22243.1 GBHO01028766 GBHO01028763 GBHO01022962 GBHO01022960 GBRD01012148 GBRD01012146 GBRD01012145 GBRD01001216 GDHC01004344 JAG14838.1 JAG14841.1 JAG20642.1 JAG20644.1 JAG53676.1 JAQ14285.1 GL450107 EFN81506.1 GDKW01000970 JAI55625.1 GBXI01015315 JAC98976.1 GDHF01007245 JAI45069.1 GAKP01008909 JAC50043.1 GAMD01002194 JAA99396.1 GAMC01002335 JAC04221.1 GFDF01006774 JAV07310.1 GECU01028182 GECU01012705 JAS79524.1 JAS95001.1 CVRI01000010 CRK89046.1 KQ435824 KOX72231.1 JXUM01098082 KQ564393 KXJ72343.1 GEBQ01024132 JAT15845.1 ABLF02028424 AK341548 BAH71918.1 GADI01006371 JAA67437.1 RCHS01002300 RMX48296.1 IACF01005695 LAB71278.1 AMQN01014763 AMQN01014764 KB311248 ELT89640.1 DS469519 EDO47515.1 NEDP02076749 OWF34844.1 GFAA01001759 JAU01676.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000037510
UP000053240
UP000075809
+ More
UP000005205 UP000027135 UP000000311 UP000007266 UP000279307 UP000007151 UP000215335 UP000002358 UP000009046 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000036403 UP000092462 UP000092461 UP000235965 UP000008820 UP000075883 UP000075900 UP000075920 UP000019118 UP000030742 UP000053097 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000242457 UP000053825 UP000192223 UP000075886 UP000075881 UP000075885 UP000005203 UP000002320 UP000015103 UP000079169 UP000008237 UP000075884 UP000076408 UP000183832 UP000053105 UP000069940 UP000249989 UP000069272 UP000007819 UP000275408 UP000014760 UP000001593 UP000242188
UP000005205 UP000027135 UP000000311 UP000007266 UP000279307 UP000007151 UP000215335 UP000002358 UP000009046 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000036403 UP000092462 UP000092461 UP000235965 UP000008820 UP000075883 UP000075900 UP000075920 UP000019118 UP000030742 UP000053097 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000242457 UP000053825 UP000192223 UP000075886 UP000075881 UP000075885 UP000005203 UP000002320 UP000015103 UP000079169 UP000008237 UP000075884 UP000076408 UP000183832 UP000053105 UP000069940 UP000249989 UP000069272 UP000007819 UP000275408 UP000014760 UP000001593 UP000242188
Pfam
PF13499 EF-hand_7
Interpro
Gene 3D
CDD
ProteinModelPortal
Q2F616
A0A2W1BJA8
A0A2A4KAH6
A0A1S6M472
I4DNU1
S4PM05
+ More
H9J942 A0A0L7LH95 A0A194R5Z1 A0A151WYP7 A0A158P2Z9 A0A067REN5 E2AUN1 A0A1B6FR13 A0A0T6B2I7 A0A1B6KFG0 A0A1Y1KNI6 A0A139WI15 V5H0Y6 A0A3L8DQV1 A0A212EJ40 A0A232F8E0 K7J3T7 E0VJM0 A0A1B6CPG4 A0A0L7K2G8 A0A0K8TS38 A0A195CEE0 A0A195DPF2 F4X5F4 A0A195BNE3 A0A195EZ36 A0A0J7L569 A0A1B0GMU7 A0A1B0GKG0 A0A0C9RSC9 A0A2H1WN62 A0A1L8DW92 A0A2J7QVV7 Q17IW0 A0A182LUX7 A0A182RRL5 A0A182WHG1 A0A023EI62 N6TET7 A0A026W121 A0A2M4C0S8 A0A182VHW9 A0A182TKX6 A0A182X145 A0A182L093 Q7Q4L8 A0A182IBY3 J3JZ87 A0A2A3EML9 A0A2M3ZJD8 A0A2M4CTB6 A0A0L7QK37 A0A1W4WK49 A0A182QZS5 A0A182K5H8 A0A182P661 A0A088ADJ0 A0A1Q3FGD1 B0W8S6 A0A2M4AUK2 U5EZE3 A0A0V0G8A0 R4WNU4 A0A023FCK3 T1HPB1 A0A336M077 A0A1S3CTK2 A0A0A9XL40 E2BRZ0 A0A0P4VSZ7 A0A1W4WJ65 A0A182NUU2 A0A0A1WJF2 A0A0K8W1T1 A0A034W8E1 A0A182Y5I2 T1DQY5 W8BS87 A0A1L8DLT9 A0A1B6HXZ2 A0A1J1HRE3 A0A0N0BEQ2 A0A182GYS5 A0A182F2B9 A0A1B6KWN8 J9K310 C4WVJ8 A0A0K8R8E5 A0A3M6U3X9 A0A2P2IB89 R7T7Q4 A7RM01 A0A210PED7 A0A1E1XQU4
H9J942 A0A0L7LH95 A0A194R5Z1 A0A151WYP7 A0A158P2Z9 A0A067REN5 E2AUN1 A0A1B6FR13 A0A0T6B2I7 A0A1B6KFG0 A0A1Y1KNI6 A0A139WI15 V5H0Y6 A0A3L8DQV1 A0A212EJ40 A0A232F8E0 K7J3T7 E0VJM0 A0A1B6CPG4 A0A0L7K2G8 A0A0K8TS38 A0A195CEE0 A0A195DPF2 F4X5F4 A0A195BNE3 A0A195EZ36 A0A0J7L569 A0A1B0GMU7 A0A1B0GKG0 A0A0C9RSC9 A0A2H1WN62 A0A1L8DW92 A0A2J7QVV7 Q17IW0 A0A182LUX7 A0A182RRL5 A0A182WHG1 A0A023EI62 N6TET7 A0A026W121 A0A2M4C0S8 A0A182VHW9 A0A182TKX6 A0A182X145 A0A182L093 Q7Q4L8 A0A182IBY3 J3JZ87 A0A2A3EML9 A0A2M3ZJD8 A0A2M4CTB6 A0A0L7QK37 A0A1W4WK49 A0A182QZS5 A0A182K5H8 A0A182P661 A0A088ADJ0 A0A1Q3FGD1 B0W8S6 A0A2M4AUK2 U5EZE3 A0A0V0G8A0 R4WNU4 A0A023FCK3 T1HPB1 A0A336M077 A0A1S3CTK2 A0A0A9XL40 E2BRZ0 A0A0P4VSZ7 A0A1W4WJ65 A0A182NUU2 A0A0A1WJF2 A0A0K8W1T1 A0A034W8E1 A0A182Y5I2 T1DQY5 W8BS87 A0A1L8DLT9 A0A1B6HXZ2 A0A1J1HRE3 A0A0N0BEQ2 A0A182GYS5 A0A182F2B9 A0A1B6KWN8 J9K310 C4WVJ8 A0A0K8R8E5 A0A3M6U3X9 A0A2P2IB89 R7T7Q4 A7RM01 A0A210PED7 A0A1E1XQU4
PDB
2LM5
E-value=2.04515e-35,
Score=369
Ontologies
GO
Topology
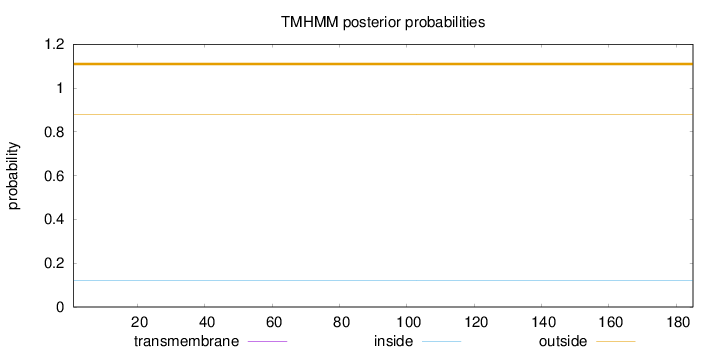
Length:
185
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12008
outside
1 - 185
Population Genetic Test Statistics
Pi
241.256332
Theta
200.841183
Tajima's D
0.649517
CLR
0.617901
CSRT
0.552122393880306
Interpretation
Uncertain
