Gene
KWMTBOMO01689
Annotation
PREDICTED:_piggyBac_transposable_element-derived_protein_4-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.456
Sequence
CDS
ATGTCCGAGCTAGATATAAACTCTGAGAACTTATTTTCACCTCAGGTTAATTTACGTGAAATCGATTTAGATAATGCATACGATTTAGAGGCGAGCGGAGATGAGATTATCCATCCCCCAAAAAAACGAAAGCGTCTTTATATAAGAAGGGATTCAGAAACTAAATGTGATTTGGTGGAAAATGTTGTCACTGCAAATGTAGCTCATGTGTCTTCAAGTAGGCTACAAAAATGGAGAGAACCAACGGAACACCAACTATCGCAGATTGCATTCACGGAGCCAACGGGTATCAAGAAACCGTATGACGAAGAACTACGAAATGCAGATCCAGGGGCCTATTTCTCATTAATAGTCTCAGACGACATTTTTGAGGAAATCGCAGTGCAAACCAACTTGTATGCTGAGCGACGCGAGAGCACAATGCCTTCCTCGCGTTTGAATAGTTGGAAGCCAACAAACAAAAATGAAGTCAAGAGTCTTTTTGGACTTATTATGTACATGGGACTAGTCAAACTCCCAAAAATAAGTCTATACTGGAGCACAGACAGGGTTTTCAAACAAACTTTTGCGCCTTCGATTATGAGTCGAAATAGATTCGAACTGTTGTTGCAGAATCTGCATTTCGTTGAAAACGAAATAGCTGATGCCGCCGATAGAATCTATAAAATACGCAGTCTGATAGATAAGTTAAACAAATCATTTCAACGGTGCTATTCGCCCAAAGAAGAAGTTTGTGTCGACGAAAGTCAAGTCCCTTACAGCGGGCGATTGATCTTAAGGCAGGGCAATAAAAGAAAAAAGCATAAATGTGGAATGAAACTGGTCAAACTGTGCACAATACCAGGTTATACCTATAAATTAGAACTTTACGCTGGTAAAAACCACGAACCACTCAATACAACGCCTAAAAATGTTGTAATGTCAATCTGTAAAGAAATACTTGATTTGGGGCACACCGTAGCTACGAATAATTCGTACACAAGTTTGGATTTAGCCCACGAACTTCTAGACAGGAATACTCATTTATTAGGCACACTGAGAAAAAATAGTAAGGGCCTCCCGAAACAAGTAATTGATGCCAAACTTAAACAAGGCGAATCTGTGGAAATGGAAAACGAAAGAGGCATAACAGTGTTACACTGGAAAGATGAACGAAGTGTGTTAATGCTCTCCACAAAACACACTAAAGAATTTCCTAGTGTGCCCATAAAAGGAAAAATGGTTCGAACACCGAAATTGATTATAGCGTATAATAAAGCAAAAGGAGCTATTCACACGAGCGATCAAATGAACGCATACTCATCACCATTGAGAAAAACTCTAAAATGGTATAAAAAGTTAGGCATCGAATTAATTTTAAACACGGCCGTAGTCAACGCCTGGATCATGTTTTGCGAAAATACGGCTCAGCAAGGAATTGTAGAATTCCGACGTCAATTAGTGGAATATCTAGAATAA
Protein
MSELDINSENLFSPQVNLREIDLDNAYDLEASGDEIIHPPKKRKRLYIRRDSETKCDLVENVVTANVAHVSSSRLQKWREPTEHQLSQIAFTEPTGIKKPYDEELRNADPGAYFSLIVSDDIFEEIAVQTNLYAERRESTMPSSRLNSWKPTNKNEVKSLFGLIMYMGLVKLPKISLYWSTDRVFKQTFAPSIMSRNRFELLLQNLHFVENEIADAADRIYKIRSLIDKLNKSFQRCYSPKEEVCVDESQVPYSGRLILRQGNKRKKHKCGMKLVKLCTIPGYTYKLELYAGKNHEPLNTTPKNVVMSICKEILDLGHTVATNNSYTSLDLAHELLDRNTHLLGTLRKNSKGLPKQVIDAKLKQGESVEMENERGITVLHWKDERSVLMLSTKHTKEFPSVPIKGKMVRTPKLIIAYNKAKGAIHTSDQMNAYSSPLRKTLKWYKKLGIELILNTAVVNAWIMFCENTAQQGIVEFRRQLVEYLE
Summary
Similarity
Belongs to the universal ribosomal protein uS3 family.
Uniprot
S4P945
A0A023EYQ2
A0A023F0B8
S4NY54
E2C9Y4
A0A084W7H9
+ More
A0A2S2QDJ8 A0A2H8TZ84 J9K6D0 X1WU42 A0A016SIZ8 J9LWR0 S4PBM2 X1X046 J9JQ12 X1XRD4 J9K106 A0A0N4XY30 J9JVX5 J9LK68 A0A1B6HG03 A0A016V127 A0A016U6U0 J9LKV2 A0A0P4WIX9 A0A0N4WU61 A0A016VFC3 A0A016VFZ3 S4P9J6 A0A0N4YQB3 J9KXH3 X1XBY4 A0A1E1WBV4 A0A023EYN9 A0A1E1VXQ8 A0A0D6LHL4 J9JPM8 T0MM39 A0A016VEZ7 A0A0P4W9M6 W6NB59 T1I3L1 A0A0N4WS15 A0A1B6IUZ3 A0A183G8F5 A0A3P8A9H6 V5GG67 A0A1E1WHA1 A0A182W9Z3 A0A016T3Q5 A0A183F4M8 A0A3P7UFA4 A0A368GTV8 A0A2B4SRV7 A0A0D8XH70 A0A2S2QUS6 J9KLM1 J9LP86 A0A087TLT3 A0A1B6K2A9 A0A1A9ZX35 A0A1B6E873 A0A0N4WGU5 A0A1A9VCE3 A0A0N4UUN0 A0A2H1V5W0 W4ZHM1 A0A1A9XAR1 A8J4K9 H9IWM5 A0A1B6LW93 A0A1B6KKZ3 A0A1B6H7X0 A0A0A9W0B8 A0A1B6LFF4 H9J421 A0A183G7U2 A0A3P8A9A8 A0A1B6J0Q3 A8J4K3 A8J4K6 A0A0A9W581
A0A2S2QDJ8 A0A2H8TZ84 J9K6D0 X1WU42 A0A016SIZ8 J9LWR0 S4PBM2 X1X046 J9JQ12 X1XRD4 J9K106 A0A0N4XY30 J9JVX5 J9LK68 A0A1B6HG03 A0A016V127 A0A016U6U0 J9LKV2 A0A0P4WIX9 A0A0N4WU61 A0A016VFC3 A0A016VFZ3 S4P9J6 A0A0N4YQB3 J9KXH3 X1XBY4 A0A1E1WBV4 A0A023EYN9 A0A1E1VXQ8 A0A0D6LHL4 J9JPM8 T0MM39 A0A016VEZ7 A0A0P4W9M6 W6NB59 T1I3L1 A0A0N4WS15 A0A1B6IUZ3 A0A183G8F5 A0A3P8A9H6 V5GG67 A0A1E1WHA1 A0A182W9Z3 A0A016T3Q5 A0A183F4M8 A0A3P7UFA4 A0A368GTV8 A0A2B4SRV7 A0A0D8XH70 A0A2S2QUS6 J9KLM1 J9LP86 A0A087TLT3 A0A1B6K2A9 A0A1A9ZX35 A0A1B6E873 A0A0N4WGU5 A0A1A9VCE3 A0A0N4UUN0 A0A2H1V5W0 W4ZHM1 A0A1A9XAR1 A8J4K9 H9IWM5 A0A1B6LW93 A0A1B6KKZ3 A0A1B6H7X0 A0A0A9W0B8 A0A1B6LFF4 H9J421 A0A183G7U2 A0A3P8A9A8 A0A1B6J0Q3 A8J4K3 A8J4K6 A0A0A9W581
Pubmed
EMBL
GAIX01005691
JAA86869.1
GBBI01004440
JAC14272.1
GBBI01004438
JAC14274.1
+ More
GAIX01013935 JAA78625.1 GL453910 EFN75248.1 ATLV01021248 KE525315 KFB46173.1 GGMS01006604 MBY75807.1 GFXV01007414 MBW19219.1 ABLF02025866 ABLF02025869 ABLF02025877 ABLF02018838 JARK01001552 EYB90668.1 ABLF02029982 GAIX01004551 JAA88009.1 ABLF02011864 ABLF02011866 ABLF02042566 ABLF02010187 ABLF02050407 ABLF02012265 ABLF02038619 UYSL01019948 VDL71548.1 ABLF02030622 ABLF02030627 ABLF02011361 GECU01034165 JAS73541.1 JARK01001356 EYC20951.1 JARK01001390 EYC10641.1 ABLF02029983 GDRN01069744 JAI63996.1 JARK01001346 EYC26339.1 EYC26340.1 GAIX01003729 JAA88831.1 UYSL01024178 VDL83170.1 ABLF02011601 ABLF02011602 ABLF02016107 GDQN01006584 JAT84470.1 GBBI01004439 JAC14273.1 GDQN01011536 GDQN01001322 JAT79518.1 JAT89732.1 KE125300 EPB69401.1 ABLF02017387 KE647018 EQB62075.1 EYC26204.1 GDRN01075668 JAI63019.1 CAVP010055412 CDL94120.1 ACPB03026393 UZAF01018515 VDO52364.1 GECU01016947 JAS90759.1 UZAH01030493 VDP10747.1 GALX01005527 JAB62939.1 GDQN01004768 JAT86286.1 JARK01001477 EYB97365.1 UZAH01001106 VDO19503.1 JOJR01000056 RCN47811.1 LSMT01000021 PFX32631.1 KN716593 KJH43099.1 GGMS01012057 MBY81260.1 ABLF02028028 ABLF02006169 ABLF02055685 KK115799 KFM66072.1 GECU01002110 JAT05597.1 GEDC01003168 JAS34130.1 UZAF01017203 VDO39079.1 UXUI01007140 VDD85673.1 ODYU01000651 SOQ35792.1 AAGJ04151167 AB300558 BAF82021.1 BABH01021160 BABH01021161 BABH01021162 BABH01021163 GEBQ01012026 JAT27951.1 GEBQ01027849 JAT12128.1 GECU01037003 GECU01013756 JAS70703.1 JAS93950.1 GBHO01042743 GDHC01013469 JAG00861.1 JAQ05160.1 GEBQ01017560 JAT22417.1 BABH01041064 UZAH01030316 VDP10112.1 GECU01014964 JAS92742.1 AB300556 BAF82019.1 AB300557 BAF82020.1 GBHO01041038 JAG02566.1
GAIX01013935 JAA78625.1 GL453910 EFN75248.1 ATLV01021248 KE525315 KFB46173.1 GGMS01006604 MBY75807.1 GFXV01007414 MBW19219.1 ABLF02025866 ABLF02025869 ABLF02025877 ABLF02018838 JARK01001552 EYB90668.1 ABLF02029982 GAIX01004551 JAA88009.1 ABLF02011864 ABLF02011866 ABLF02042566 ABLF02010187 ABLF02050407 ABLF02012265 ABLF02038619 UYSL01019948 VDL71548.1 ABLF02030622 ABLF02030627 ABLF02011361 GECU01034165 JAS73541.1 JARK01001356 EYC20951.1 JARK01001390 EYC10641.1 ABLF02029983 GDRN01069744 JAI63996.1 JARK01001346 EYC26339.1 EYC26340.1 GAIX01003729 JAA88831.1 UYSL01024178 VDL83170.1 ABLF02011601 ABLF02011602 ABLF02016107 GDQN01006584 JAT84470.1 GBBI01004439 JAC14273.1 GDQN01011536 GDQN01001322 JAT79518.1 JAT89732.1 KE125300 EPB69401.1 ABLF02017387 KE647018 EQB62075.1 EYC26204.1 GDRN01075668 JAI63019.1 CAVP010055412 CDL94120.1 ACPB03026393 UZAF01018515 VDO52364.1 GECU01016947 JAS90759.1 UZAH01030493 VDP10747.1 GALX01005527 JAB62939.1 GDQN01004768 JAT86286.1 JARK01001477 EYB97365.1 UZAH01001106 VDO19503.1 JOJR01000056 RCN47811.1 LSMT01000021 PFX32631.1 KN716593 KJH43099.1 GGMS01012057 MBY81260.1 ABLF02028028 ABLF02006169 ABLF02055685 KK115799 KFM66072.1 GECU01002110 JAT05597.1 GEDC01003168 JAS34130.1 UZAF01017203 VDO39079.1 UXUI01007140 VDD85673.1 ODYU01000651 SOQ35792.1 AAGJ04151167 AB300558 BAF82021.1 BABH01021160 BABH01021161 BABH01021162 BABH01021163 GEBQ01012026 JAT27951.1 GEBQ01027849 JAT12128.1 GECU01037003 GECU01013756 JAS70703.1 JAS93950.1 GBHO01042743 GDHC01013469 JAG00861.1 JAQ05160.1 GEBQ01017560 JAT22417.1 BABH01041064 UZAH01030316 VDP10112.1 GECU01014964 JAS92742.1 AB300556 BAF82019.1 AB300557 BAF82020.1 GBHO01041038 JAG02566.1
Proteomes
Pfam
Interpro
IPR029526
PGBD
+ More
IPR008906 HATC_C_dom
IPR012337 RNaseH-like_sf
IPR003034 SAP_dom
IPR036361 SAP_dom_sf
IPR008042 Retrotrans_Pao
IPR014044 CAP_domain
IPR035940 CAP_sf
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR001351 Ribosomal_S3_C
IPR004044 KH_dom_type_2
IPR018280 Ribosomal_S3_CS
IPR015946 KH_dom-like_a/b
IPR009019 KH_sf_prok-type
IPR032718 PGBD4_C_Znf-ribbon
IPR000315 Znf_B-box
IPR007889 HTH_Psq
IPR009057 Homeobox-like_sf
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR008906 HATC_C_dom
IPR012337 RNaseH-like_sf
IPR003034 SAP_dom
IPR036361 SAP_dom_sf
IPR008042 Retrotrans_Pao
IPR014044 CAP_domain
IPR035940 CAP_sf
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR001351 Ribosomal_S3_C
IPR004044 KH_dom_type_2
IPR018280 Ribosomal_S3_CS
IPR015946 KH_dom-like_a/b
IPR009019 KH_sf_prok-type
IPR032718 PGBD4_C_Znf-ribbon
IPR000315 Znf_B-box
IPR007889 HTH_Psq
IPR009057 Homeobox-like_sf
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
SUPFAM
Gene 3D
ProteinModelPortal
S4P945
A0A023EYQ2
A0A023F0B8
S4NY54
E2C9Y4
A0A084W7H9
+ More
A0A2S2QDJ8 A0A2H8TZ84 J9K6D0 X1WU42 A0A016SIZ8 J9LWR0 S4PBM2 X1X046 J9JQ12 X1XRD4 J9K106 A0A0N4XY30 J9JVX5 J9LK68 A0A1B6HG03 A0A016V127 A0A016U6U0 J9LKV2 A0A0P4WIX9 A0A0N4WU61 A0A016VFC3 A0A016VFZ3 S4P9J6 A0A0N4YQB3 J9KXH3 X1XBY4 A0A1E1WBV4 A0A023EYN9 A0A1E1VXQ8 A0A0D6LHL4 J9JPM8 T0MM39 A0A016VEZ7 A0A0P4W9M6 W6NB59 T1I3L1 A0A0N4WS15 A0A1B6IUZ3 A0A183G8F5 A0A3P8A9H6 V5GG67 A0A1E1WHA1 A0A182W9Z3 A0A016T3Q5 A0A183F4M8 A0A3P7UFA4 A0A368GTV8 A0A2B4SRV7 A0A0D8XH70 A0A2S2QUS6 J9KLM1 J9LP86 A0A087TLT3 A0A1B6K2A9 A0A1A9ZX35 A0A1B6E873 A0A0N4WGU5 A0A1A9VCE3 A0A0N4UUN0 A0A2H1V5W0 W4ZHM1 A0A1A9XAR1 A8J4K9 H9IWM5 A0A1B6LW93 A0A1B6KKZ3 A0A1B6H7X0 A0A0A9W0B8 A0A1B6LFF4 H9J421 A0A183G7U2 A0A3P8A9A8 A0A1B6J0Q3 A8J4K3 A8J4K6 A0A0A9W581
A0A2S2QDJ8 A0A2H8TZ84 J9K6D0 X1WU42 A0A016SIZ8 J9LWR0 S4PBM2 X1X046 J9JQ12 X1XRD4 J9K106 A0A0N4XY30 J9JVX5 J9LK68 A0A1B6HG03 A0A016V127 A0A016U6U0 J9LKV2 A0A0P4WIX9 A0A0N4WU61 A0A016VFC3 A0A016VFZ3 S4P9J6 A0A0N4YQB3 J9KXH3 X1XBY4 A0A1E1WBV4 A0A023EYN9 A0A1E1VXQ8 A0A0D6LHL4 J9JPM8 T0MM39 A0A016VEZ7 A0A0P4W9M6 W6NB59 T1I3L1 A0A0N4WS15 A0A1B6IUZ3 A0A183G8F5 A0A3P8A9H6 V5GG67 A0A1E1WHA1 A0A182W9Z3 A0A016T3Q5 A0A183F4M8 A0A3P7UFA4 A0A368GTV8 A0A2B4SRV7 A0A0D8XH70 A0A2S2QUS6 J9KLM1 J9LP86 A0A087TLT3 A0A1B6K2A9 A0A1A9ZX35 A0A1B6E873 A0A0N4WGU5 A0A1A9VCE3 A0A0N4UUN0 A0A2H1V5W0 W4ZHM1 A0A1A9XAR1 A8J4K9 H9IWM5 A0A1B6LW93 A0A1B6KKZ3 A0A1B6H7X0 A0A0A9W0B8 A0A1B6LFF4 H9J421 A0A183G7U2 A0A3P8A9A8 A0A1B6J0Q3 A8J4K3 A8J4K6 A0A0A9W581
Ontologies
KEGG
GO
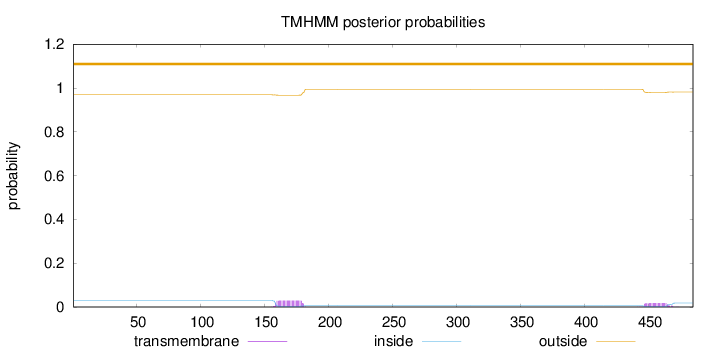
Topology
Subcellular location
Nucleus
Length:
485
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00582
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03117
outside
1 - 485
Population Genetic Test Statistics
Pi
370.510352
Theta
235.012597
Tajima's D
1.978067
CLR
0
CSRT
0.879606019699015
Interpretation
Uncertain
