Gene
KWMTBOMO01686
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101744003_[Bombyx_mori]
Full name
Tetraspanin
+ More
Mitogen-activated protein kinase kinase kinase kinase
Mitogen-activated protein kinase kinase kinase kinase
Location in the cell
Nuclear Reliability : 3.843
Sequence
CDS
ATGTGGGCCGGCATTTCATTCGACGGTAAAACCGAGCTTGTTTGCGTGCCTGGCGGGGGACAAGGCGGCGGTCTAACTTCGGACCGGTACATTTCCGATATTCTGCTGGAACATGTCGTTCCCTATGCGGGATATATCGGTGATGCCTTGCTCCTTATCCACGATAAAGCTCGTTGTCACACTGCCCGTGTAACAACTGAATACCTCGAAGAAGTCGGTATCGCCACATTGGACTGGCCAGCGCTCAGCTTGGACTTGAATCCCATTGAGCACGTGTGGGATGAACTGAAGAGGAAGGTTCGTTCCAGAACTCCTGCTCTTTCATGTCTGAATGAGCTGAAATCTACGTTGATTGAGAAGTGGGAAGGTATCCCACAAGAATCAATTCAGAAGTTGATCAGGTCTATGAAGAATCGTCTTCGAGCAGTTATTGGGGCGAGGGGAGGGAACACCAAGTACTGA
Protein
MWAGISFDGKTELVCVPGGGQGGGLTSDRYISDILLEHVVPYAGYIGDALLLIHDKARCHTARVTTEYLEEVGIATLDWPALSLDLNPIEHVWDELKRKVRSRTPALSCLNELKSTLIEKWEGIPQESIQKLIRSMKNRLRAVIGARGGNTKY
Summary
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Similarity
Belongs to the tetraspanin (TM4SF) family.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. STE20 subfamily.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. STE20 subfamily.
Uniprot
A0A2H1W7Z9
A0A2W1BPI3
A0A2W1C099
A0A2P8XLM9
A0A060ZHS7
V5G1E7
+ More
A0A1A7WG79 A0A060WX74 A0A3B5QB47 A0A3B5QQ76 A0A3B5R2Z8 A0A3B3C4K6 A0A3B3BTS0 A0A3B3BLR3 A0A2W1BJ04 A0A3B3CHP7 A0A060ZBB1 A0A1A7YAS8 A0A3B3BAG0 A0A3B3CSK6 A0A3B5QT88 A0A3B3C7G6 A0A3P9NLT4 U3FVR2 A0A3B3BAD7 A0A3P9QFR2 A0A060VRW1 A0A060X227 A0A060WIG2 A0A060XIF3 A0A3P9QB88 A0A3B5REA1 A0A060WNH7 A8C5I4 A0A060XHZ6 A0A060YT72 A0A060Z1P8 A0A060YS83 A0A3B5PR17 A0A3B3B9N3 A0A3B3DMT7 U4U8G8 A0A3B4CPA3 A0A1A7WB56 A0A060YFQ7 A0A3B5QE26 A0A060W3F6 A0A3B4DMX9 A0A3P8U4P3 A0A060WZK2 A0A060YJG4 A0A060WZ07 A0A061ADR3 A0A3B3BMS0 A0A060Z353 A0A3B3D6L6 A0A3B5B4E9 A0A060WG12 A0A060ZEG9 A0A060ZAN4 A0A2D4P6Z1 A0A0J7N2U1 A0A060X263 A0A3B5QMY4 A0A060X3S3 R4FJ13 A0A3B3RC03 A0A3B5A2U8 A0A3P9P6G5 A0A060YTF8 A0A3B5KAA3 D2A2Q9 A0A2A4J5V2 A0A087TI70 A0A060ZGV7 A0A3P9PTW2 A0A060XQ36 A0A060YH30 A0A3B3QX33 A0A3B3QA75 A0A3P8YCF0 A0A087TJI5 A0A139W8R7 A0A3B4VAJ1 A0A060XL34 A0A067RM16 A0A0J7JXZ6 A0A3P9HRI8 A0A3B5BL52 A0A3P8RUJ6 A0A3B3CYT8 A0A3P9JG58 A0A3B4DRB4 A0A023F6N5 A0A3N0Z9J3 A0A060XFH1 A0A060VWW5 A0A3P9HL43
A0A1A7WG79 A0A060WX74 A0A3B5QB47 A0A3B5QQ76 A0A3B5R2Z8 A0A3B3C4K6 A0A3B3BTS0 A0A3B3BLR3 A0A2W1BJ04 A0A3B3CHP7 A0A060ZBB1 A0A1A7YAS8 A0A3B3BAG0 A0A3B3CSK6 A0A3B5QT88 A0A3B3C7G6 A0A3P9NLT4 U3FVR2 A0A3B3BAD7 A0A3P9QFR2 A0A060VRW1 A0A060X227 A0A060WIG2 A0A060XIF3 A0A3P9QB88 A0A3B5REA1 A0A060WNH7 A8C5I4 A0A060XHZ6 A0A060YT72 A0A060Z1P8 A0A060YS83 A0A3B5PR17 A0A3B3B9N3 A0A3B3DMT7 U4U8G8 A0A3B4CPA3 A0A1A7WB56 A0A060YFQ7 A0A3B5QE26 A0A060W3F6 A0A3B4DMX9 A0A3P8U4P3 A0A060WZK2 A0A060YJG4 A0A060WZ07 A0A061ADR3 A0A3B3BMS0 A0A060Z353 A0A3B3D6L6 A0A3B5B4E9 A0A060WG12 A0A060ZEG9 A0A060ZAN4 A0A2D4P6Z1 A0A0J7N2U1 A0A060X263 A0A3B5QMY4 A0A060X3S3 R4FJ13 A0A3B3RC03 A0A3B5A2U8 A0A3P9P6G5 A0A060YTF8 A0A3B5KAA3 D2A2Q9 A0A2A4J5V2 A0A087TI70 A0A060ZGV7 A0A3P9PTW2 A0A060XQ36 A0A060YH30 A0A3B3QX33 A0A3B3QA75 A0A3P8YCF0 A0A087TJI5 A0A139W8R7 A0A3B4VAJ1 A0A060XL34 A0A067RM16 A0A0J7JXZ6 A0A3P9HRI8 A0A3B5BL52 A0A3P8RUJ6 A0A3B3CYT8 A0A3P9JG58 A0A3B4DRB4 A0A023F6N5 A0A3N0Z9J3 A0A060XFH1 A0A060VWW5 A0A3P9HL43
EC Number
2.7.11.1
Pubmed
EMBL
ODYU01006928
SOQ49209.1
KZ150035
PZC74640.1
KZ149928
PZC77493.1
+ More
PYGN01001767 PSN32920.1 FR965136 CDR01129.1 GALX01004626 JAB63840.1 HADW01003134 SBP04534.1 FR904804 CDQ71978.1 KZ150024 PZC74838.1 FR938374 CDQ98570.1 HADX01005375 HADX01006576 SBP27607.1 SBP28808.1 GAEP01001740 JAB53081.1 FR904289 CDQ57597.1 FR904917 CDQ73648.1 FR904491 CDQ65039.1 FR905160 CDQ76660.1 FR904637 CDQ68666.1 EF685955 ABV31711.1 FR905186 CDQ76934.1 FR913091 CDQ92325.1 FR934058 CDQ97956.1 FR918129 CDQ94591.1 KB632087 ERL88653.1 HADW01001545 SBP02945.1 FR910704 CDQ90586.1 FR904389 CDQ61793.1 FR904832 CDQ72427.1 FR912605 CDQ92013.1 FR904707 CDQ70284.1 FR979969 CDR18852.1 FR924350 CDQ96159.1 FR904534 CDQ66203.1 FR948558 CDQ99615.1 FR939895 CDQ98734.1 IACN01051383 LAB53764.1 LBMM01011147 KMQ87000.1 FR904893 CDQ73307.1 CDQ71979.1 GAHY01002463 JAA75047.1 FR918761 CDQ94842.1 KQ971339 EFA02765.1 NWSH01003130 PCG66904.1 KK115333 KFM64809.1 FR961204 CDR00794.1 FR905754 CDQ81397.1 FR910992 CDQ90832.1 KK115499 KFM65274.1 KQ973073 KXZ75680.1 FR905246 CDQ77605.1 KK852595 KDR20647.1 LBMM01023366 KMQ82736.1 GBBI01001820 JAC16892.1 RJVU01002272 ROL55109.1 FR905317 CDQ78348.1 FR904320 CDQ59271.1
PYGN01001767 PSN32920.1 FR965136 CDR01129.1 GALX01004626 JAB63840.1 HADW01003134 SBP04534.1 FR904804 CDQ71978.1 KZ150024 PZC74838.1 FR938374 CDQ98570.1 HADX01005375 HADX01006576 SBP27607.1 SBP28808.1 GAEP01001740 JAB53081.1 FR904289 CDQ57597.1 FR904917 CDQ73648.1 FR904491 CDQ65039.1 FR905160 CDQ76660.1 FR904637 CDQ68666.1 EF685955 ABV31711.1 FR905186 CDQ76934.1 FR913091 CDQ92325.1 FR934058 CDQ97956.1 FR918129 CDQ94591.1 KB632087 ERL88653.1 HADW01001545 SBP02945.1 FR910704 CDQ90586.1 FR904389 CDQ61793.1 FR904832 CDQ72427.1 FR912605 CDQ92013.1 FR904707 CDQ70284.1 FR979969 CDR18852.1 FR924350 CDQ96159.1 FR904534 CDQ66203.1 FR948558 CDQ99615.1 FR939895 CDQ98734.1 IACN01051383 LAB53764.1 LBMM01011147 KMQ87000.1 FR904893 CDQ73307.1 CDQ71979.1 GAHY01002463 JAA75047.1 FR918761 CDQ94842.1 KQ971339 EFA02765.1 NWSH01003130 PCG66904.1 KK115333 KFM64809.1 FR961204 CDR00794.1 FR905754 CDQ81397.1 FR910992 CDQ90832.1 KK115499 KFM65274.1 KQ973073 KXZ75680.1 FR905246 CDQ77605.1 KK852595 KDR20647.1 LBMM01023366 KMQ82736.1 GBBI01001820 JAC16892.1 RJVU01002272 ROL55109.1 FR905317 CDQ78348.1 FR904320 CDQ59271.1
Proteomes
Pfam
Interpro
IPR002492
Transposase_Tc1-like
+ More
IPR009057 Homeobox-like_sf
IPR038717 Tc1-like_DDE_dom
IPR036388 WH-like_DNA-bd_sf
IPR001523 Paired_dom
IPR012337 RNaseH-like_sf
IPR000301 Tetraspanin
IPR018499 Tetraspanin/Peripherin
IPR008952 Tetraspanin_EC2_sf
IPR006579 Pre_C2HC_dom
IPR001878 Znf_CCHC
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR000719 Prot_kinase_dom
IPR021160 MAPKKKK
IPR001180 CNH_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR036179 Ig-like_dom_sf
IPR013106 Ig_V-set
IPR015714 Lymphocyte_func-assoc_Ag_3
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR009057 Homeobox-like_sf
IPR038717 Tc1-like_DDE_dom
IPR036388 WH-like_DNA-bd_sf
IPR001523 Paired_dom
IPR012337 RNaseH-like_sf
IPR000301 Tetraspanin
IPR018499 Tetraspanin/Peripherin
IPR008952 Tetraspanin_EC2_sf
IPR006579 Pre_C2HC_dom
IPR001878 Znf_CCHC
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR000719 Prot_kinase_dom
IPR021160 MAPKKKK
IPR001180 CNH_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR036179 Ig-like_dom_sf
IPR013106 Ig_V-set
IPR015714 Lymphocyte_func-assoc_Ag_3
IPR013783 Ig-like_fold
IPR003599 Ig_sub
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1W7Z9
A0A2W1BPI3
A0A2W1C099
A0A2P8XLM9
A0A060ZHS7
V5G1E7
+ More
A0A1A7WG79 A0A060WX74 A0A3B5QB47 A0A3B5QQ76 A0A3B5R2Z8 A0A3B3C4K6 A0A3B3BTS0 A0A3B3BLR3 A0A2W1BJ04 A0A3B3CHP7 A0A060ZBB1 A0A1A7YAS8 A0A3B3BAG0 A0A3B3CSK6 A0A3B5QT88 A0A3B3C7G6 A0A3P9NLT4 U3FVR2 A0A3B3BAD7 A0A3P9QFR2 A0A060VRW1 A0A060X227 A0A060WIG2 A0A060XIF3 A0A3P9QB88 A0A3B5REA1 A0A060WNH7 A8C5I4 A0A060XHZ6 A0A060YT72 A0A060Z1P8 A0A060YS83 A0A3B5PR17 A0A3B3B9N3 A0A3B3DMT7 U4U8G8 A0A3B4CPA3 A0A1A7WB56 A0A060YFQ7 A0A3B5QE26 A0A060W3F6 A0A3B4DMX9 A0A3P8U4P3 A0A060WZK2 A0A060YJG4 A0A060WZ07 A0A061ADR3 A0A3B3BMS0 A0A060Z353 A0A3B3D6L6 A0A3B5B4E9 A0A060WG12 A0A060ZEG9 A0A060ZAN4 A0A2D4P6Z1 A0A0J7N2U1 A0A060X263 A0A3B5QMY4 A0A060X3S3 R4FJ13 A0A3B3RC03 A0A3B5A2U8 A0A3P9P6G5 A0A060YTF8 A0A3B5KAA3 D2A2Q9 A0A2A4J5V2 A0A087TI70 A0A060ZGV7 A0A3P9PTW2 A0A060XQ36 A0A060YH30 A0A3B3QX33 A0A3B3QA75 A0A3P8YCF0 A0A087TJI5 A0A139W8R7 A0A3B4VAJ1 A0A060XL34 A0A067RM16 A0A0J7JXZ6 A0A3P9HRI8 A0A3B5BL52 A0A3P8RUJ6 A0A3B3CYT8 A0A3P9JG58 A0A3B4DRB4 A0A023F6N5 A0A3N0Z9J3 A0A060XFH1 A0A060VWW5 A0A3P9HL43
A0A1A7WG79 A0A060WX74 A0A3B5QB47 A0A3B5QQ76 A0A3B5R2Z8 A0A3B3C4K6 A0A3B3BTS0 A0A3B3BLR3 A0A2W1BJ04 A0A3B3CHP7 A0A060ZBB1 A0A1A7YAS8 A0A3B3BAG0 A0A3B3CSK6 A0A3B5QT88 A0A3B3C7G6 A0A3P9NLT4 U3FVR2 A0A3B3BAD7 A0A3P9QFR2 A0A060VRW1 A0A060X227 A0A060WIG2 A0A060XIF3 A0A3P9QB88 A0A3B5REA1 A0A060WNH7 A8C5I4 A0A060XHZ6 A0A060YT72 A0A060Z1P8 A0A060YS83 A0A3B5PR17 A0A3B3B9N3 A0A3B3DMT7 U4U8G8 A0A3B4CPA3 A0A1A7WB56 A0A060YFQ7 A0A3B5QE26 A0A060W3F6 A0A3B4DMX9 A0A3P8U4P3 A0A060WZK2 A0A060YJG4 A0A060WZ07 A0A061ADR3 A0A3B3BMS0 A0A060Z353 A0A3B3D6L6 A0A3B5B4E9 A0A060WG12 A0A060ZEG9 A0A060ZAN4 A0A2D4P6Z1 A0A0J7N2U1 A0A060X263 A0A3B5QMY4 A0A060X3S3 R4FJ13 A0A3B3RC03 A0A3B5A2U8 A0A3P9P6G5 A0A060YTF8 A0A3B5KAA3 D2A2Q9 A0A2A4J5V2 A0A087TI70 A0A060ZGV7 A0A3P9PTW2 A0A060XQ36 A0A060YH30 A0A3B3QX33 A0A3B3QA75 A0A3P8YCF0 A0A087TJI5 A0A139W8R7 A0A3B4VAJ1 A0A060XL34 A0A067RM16 A0A0J7JXZ6 A0A3P9HRI8 A0A3B5BL52 A0A3P8RUJ6 A0A3B3CYT8 A0A3P9JG58 A0A3B4DRB4 A0A023F6N5 A0A3N0Z9J3 A0A060XFH1 A0A060VWW5 A0A3P9HL43
PDB
5CR4
E-value=1.29931e-13,
Score=179
Ontologies
GO
PANTHER
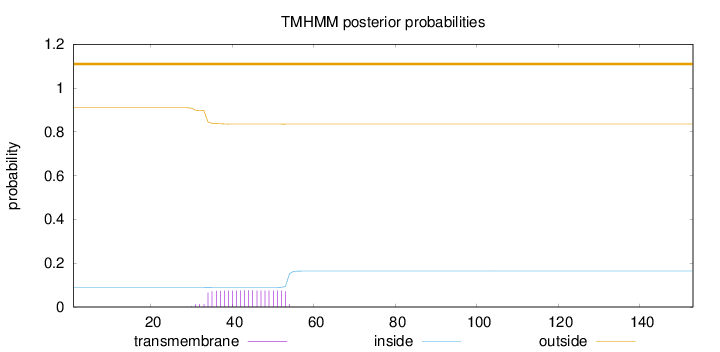
Topology
Subcellular location
Membrane
Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.54099
Exp number, first 60 AAs:
1.5408
Total prob of N-in:
0.08995
outside
1 - 153
Population Genetic Test Statistics
Pi
38.879963
Theta
49.732165
Tajima's D
1.401582
CLR
0
CSRT
0.765161741912904
Interpretation
Uncertain
