Gene
KWMTBOMO01685
Pre Gene Modal
BGIBMGA006036
Annotation
PREDICTED:_SUN_domain-containing_ossification_factor_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.014
Sequence
CDS
ATGTCGCCGCCGCGGGCGCTGCTTGCGGCTCTGCTAATATGTCAGCTGTTGTCGTACGGTGGCCACCAGGGACTGACCAAGATATTTTACACATTACCTGACACAGGGCAGCCTCCGGTTCTTATATCGTTCGGGGATAACGCGAAATTGGACCTGGGCAATACGGATGAACTAATGTTCGTAAATGACTCCGACGAGCACAACGACACCAAGGCATTTTCAATCGTCGACACCGATACAATAAAGGTTGAAAGTGAAACAGACAGTTTAGAGGACAAGCCAAGCACAGAACCGAGACTGGTTGTGAAAGCGGTCCCCAAACATGAACTGAAACAAACAGAAGATGCATCTCAACCAGACGGAGAAGTCTTAGAATCTCAAGTTGAAGAACAAATAGATGAAGAACCGGTTCACGTTCCAGTCAAACCGGAAACCCCGCAAGAAGACATCCCCTCGTTTTCGGAGTGGGCGCAGAAACAACTCGCCGAGGCAGAGAAACAGGACACAGTACTGAATCACTCGAGTCAACCTAGTCATAGTAATACTAATTTTAGTAGTAAGAGCGTGAAGTTGCGGTCGAAGAACTACGCGTCGCTGGCGTGCGGCGCCAAGGTCGTCGCCGTCAACCCGGAGGCGGGCTCGCCCAGCTCCATCCTCTCACCCAACCGGGACGAGTACATGCTCAACACGTGCAACAGTCGCATTTGGTTCGTCGTCGAACTCTGCGAAGCTGTTCAAGCTCAAAAAATAGAGATAGCCAACTTCGAGTTGTTTTCTTCAACTCCCAAAGACATCGCCGTTTACTTTAGCGATCGGTTCCCGACGAGAGACTGGGCTAGCGTCGGACAGTTCACCGCCCAAGACATGAGAGACGTGCAAAGCTTCGACCTGTACCCACATCTATTCGGCAAGTTCATAAAAGTCGAGATGCTTTCGCATCACGGCTCGGAGCATTACTGTCCGATTTCGCTGTTCAAAGTGTACGGCACTTCAGAGTTCGAGGTGCTAGAGAAGGAGAATTCGCAACATCCCGCTCATATCGACGAGGACGAAGACGACGAGATAATAGACGTTCCCGACGTGCCGGCCGCCGAGACGGAGCCCTCCAAGAACCTGTTCGGGTCCGCGCGGGACGCCGTCATGTCCATTATGAAGAAGGCGGCCCAGGCGTTAGTCAAAACGGAAGTTCCTAAAAACGTGTCGTCCGAACACAACGACACGTTGGCAGACGACGCGTACAGACGCTGTTGCTCTCCCAGCCATATTATAGTGTGTGATAATTGCAGTGAGTCGCTGTACAATGACGTATACGAACTCATTAGTTGTAACTCGGACAAATTAGTTAGTTTAATGCGTCAAGGATTCCTCCGCGATACCTTGAAGTGTACTGGCATCTGCCAACAGTTCGGTTTAGATTTTAAAAGTACAAAGACAATAGAGTTTAGTGATGAGCGTGTCGCTTACATGAACGCGTTATTCCCGCAAAAGTATTTAGCCGCCTTGTGTAATATATTAGCGATAAAAGAGAAGAAGGTCGTTTTGAATACGAGCTTTGAAAACGAACATAACGTTACTAGTGATAATAGTACTGAAGAGTCGCCGCAGGTCGTGAATAGCGGAACTAACGGCGAACAAGAGATCAAGTTGGCTCCCGACAATAGCAACGGCTCCGATGATAAAATCAATGAAACAACGCAAAGTGATGAGATACCAATTGATACGCTCAACGATAGCAGCACGTTACCTGTGGAACTTCTCCCAGAAGAAGTTGACAACATGGAGGATAAGAAAGAAGACGTTATCGTCGCAGAGGAGACTTCAGGAGATGAACCCCAAGAATTTATAGCACCAAATATTGATAAATCTATAGAAATAGCGACGGAGCAAGAAAACAAAGATGGACTTTTGAAAACAAAAATTGAAAACGGCAAGGAAATCACAGAGAAGAGAGACGGCAATGGCGAGGAGTCGAATGATCAGCTGATGATGGAGAATGACAATTTCATTTCGGATATCGATCAGATAGCCGCGGACCCCGCGCCGCCCGGCGCTCCCAACACGCAGAACCAAAATCAGCAGCAGACCACTCTGCAGAAGGAATCTGTTTTTTTGCGTCTCTCTAACAGAGTAAAGACCCTAGAACGCAACATGTCTCTATCGGGACAATACTTAGAAGAGTTGAGCAGGAGGTACAAGAGACAAGTGGAAGAGATGCAGAAGACTTTCGAGAAGACTGTCCAGCAGATGACAGAGGAGAAGAAGAAAACAAATGAACGAGAACAGAAATACTTGGAACAAATGAGCAATCTCCAGGAACAGCTGGCCCAGATGACGTCAGCAATGCACGTACTCATGGAAGAGAGAGACAGCTGGTTCGGAAACATTAACTTCTTCAGATTCATCATCTTCCAAGCTATCATTGTGGCTCTAGTTATATACTACGTCTCGAAACGAAGAAGAATCGAACCGATATTAGTTCCGGTTCCGAGAAAAACAAGAAAGAAACAGGACAAGTTGAGGCGGAAATCTGTCGAAGGGGTCAGCGGACACGCCACGCCTTCCACCAAGAAACGGAGACCCAGCGAGGAGGCTCTGCAGATAGCCAGACAAGCCATTGAGGACACCAAGGGCAGCAACGAAGGAGAGTGGCAGGTGGCTCGTAAGAACAGAAGGAGAAAAACTTGTATAGTACTTAACGCGGAGACAGCAGCCAAGAGTTGGACGCGGCAGGACAGCATCGGGAAGCTGCAGGAGAACACCATAACCTTGGATGATGACGAGTACGTGGCGCCCGTGTCCGAGCCCAAGCAGTTTAACGCGGACGTGGAGCCGCCCAAACCGGACTACACCAAGACGAATGGCTTCTTCAACAACTTGAAAACAAAAACCATGAAGACCAGGAGACTGTCATCACCTGCATTTTTGCGAACGTTCAGCCGCCAGAGCACCAGGAGCACGCCCAGCCCCGTCGTGAGGAGTGAGGAGCCGATCTTCAACGGGAACGCAAAGAAGAAGGCCGCCTCAGAGTCACCCACCGGGAGTCTCTGGTCGGAGTCCACGGAGTTGTCCCCGCAGGACAGCGAGAGCAGTGGCAGTAAAAAAAAGAAAAGTCTGAAGAATATATTGAAGAAAGTGTTTTAA
Protein
MSPPRALLAALLICQLLSYGGHQGLTKIFYTLPDTGQPPVLISFGDNAKLDLGNTDELMFVNDSDEHNDTKAFSIVDTDTIKVESETDSLEDKPSTEPRLVVKAVPKHELKQTEDASQPDGEVLESQVEEQIDEEPVHVPVKPETPQEDIPSFSEWAQKQLAEAEKQDTVLNHSSQPSHSNTNFSSKSVKLRSKNYASLACGAKVVAVNPEAGSPSSILSPNRDEYMLNTCNSRIWFVVELCEAVQAQKIEIANFELFSSTPKDIAVYFSDRFPTRDWASVGQFTAQDMRDVQSFDLYPHLFGKFIKVEMLSHHGSEHYCPISLFKVYGTSEFEVLEKENSQHPAHIDEDEDDEIIDVPDVPAAETEPSKNLFGSARDAVMSIMKKAAQALVKTEVPKNVSSEHNDTLADDAYRRCCSPSHIIVCDNCSESLYNDVYELISCNSDKLVSLMRQGFLRDTLKCTGICQQFGLDFKSTKTIEFSDERVAYMNALFPQKYLAALCNILAIKEKKVVLNTSFENEHNVTSDNSTEESPQVVNSGTNGEQEIKLAPDNSNGSDDKINETTQSDEIPIDTLNDSSTLPVELLPEEVDNMEDKKEDVIVAEETSGDEPQEFIAPNIDKSIEIATEQENKDGLLKTKIENGKEITEKRDGNGEESNDQLMMENDNFISDIDQIAADPAPPGAPNTQNQNQQQTTLQKESVFLRLSNRVKTLERNMSLSGQYLEELSRRYKRQVEEMQKTFEKTVQQMTEEKKKTNEREQKYLEQMSNLQEQLAQMTSAMHVLMEERDSWFGNINFFRFIIFQAIIVALVIYYVSKRRRIEPILVPVPRKTRKKQDKLRRKSVEGVSGHATPSTKKRRPSEEALQIARQAIEDTKGSNEGEWQVARKNRRRKTCIVLNAETAAKSWTRQDSIGKLQENTITLDDDEYVAPVSEPKQFNADVEPPKPDYTKTNGFFNNLKTKTMKTRRLSSPAFLRTFSRQSTRSTPSPVVRSEEPIFNGNAKKKAASESPTGSLWSESTELSPQDSESSGSKKKKSLKNILKKVF
Summary
Uniprot
H9J944
A0A2H1WNB8
A0A2W1BRK7
A0A2A4KB70
A0A194Q0J1
A0A0L7L992
+ More
A0A194RBG4 A0A212EJ41 A0A139WHM5 A0A139WHM0 E2BQP7 A0A0J7L2Y7 A0A1Y1LXV7 A0A1B6E7E5 A0A1B6BXW7 A0A1B6DYA5 A0A1B6E2D1 A0A0N0U7F7 A0A310SN20 A0A2R7VPZ1 A0A023EYJ1 A0A146L160 A0A146MGE9 T1HPP7 A0A146LWK8 A0A2S2R5X9 A0A336LJL6 A0A336LNX1
A0A194RBG4 A0A212EJ41 A0A139WHM5 A0A139WHM0 E2BQP7 A0A0J7L2Y7 A0A1Y1LXV7 A0A1B6E7E5 A0A1B6BXW7 A0A1B6DYA5 A0A1B6E2D1 A0A0N0U7F7 A0A310SN20 A0A2R7VPZ1 A0A023EYJ1 A0A146L160 A0A146MGE9 T1HPP7 A0A146LWK8 A0A2S2R5X9 A0A336LJL6 A0A336LNX1
Pubmed
EMBL
BABH01004457
ODYU01009823
SOQ54507.1
KZ150052
PZC74393.1
NWSH01000011
+ More
PCG80912.1 KQ459584 KPI98514.1 JTDY01002109 KOB72083.1 KQ460644 KPJ13241.1 AGBW02014536 OWR41491.1 KQ971343 KYB27462.1 KYB27463.1 GL449798 EFN81966.1 LBMM01001165 KMQ96794.1 GEZM01044286 JAV78329.1 GEDC01003524 JAS33774.1 GEDC01031187 JAS06111.1 GEDC01026899 GEDC01017845 GEDC01006644 JAS10399.1 JAS19453.1 JAS30654.1 GEDC01005203 JAS32095.1 KQ435711 KOX79424.1 KQ759840 OAD62645.1 KK854016 PTY09419.1 GBBI01004530 JAC14182.1 GDHC01017647 JAQ00982.1 GDHC01000793 JAQ17836.1 ACPB03001465 GDHC01006558 JAQ12071.1 GGMS01016264 MBY85467.1 UFQS01000037 UFQT01000037 SSW97988.1 SSX18374.1 SSW97987.1 SSX18373.1
PCG80912.1 KQ459584 KPI98514.1 JTDY01002109 KOB72083.1 KQ460644 KPJ13241.1 AGBW02014536 OWR41491.1 KQ971343 KYB27462.1 KYB27463.1 GL449798 EFN81966.1 LBMM01001165 KMQ96794.1 GEZM01044286 JAV78329.1 GEDC01003524 JAS33774.1 GEDC01031187 JAS06111.1 GEDC01026899 GEDC01017845 GEDC01006644 JAS10399.1 JAS19453.1 JAS30654.1 GEDC01005203 JAS32095.1 KQ435711 KOX79424.1 KQ759840 OAD62645.1 KK854016 PTY09419.1 GBBI01004530 JAC14182.1 GDHC01017647 JAQ00982.1 GDHC01000793 JAQ17836.1 ACPB03001465 GDHC01006558 JAQ12071.1 GGMS01016264 MBY85467.1 UFQS01000037 UFQT01000037 SSW97988.1 SSX18374.1 SSW97987.1 SSX18373.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF49785
SSF49785
Gene 3D
ProteinModelPortal
H9J944
A0A2H1WNB8
A0A2W1BRK7
A0A2A4KB70
A0A194Q0J1
A0A0L7L992
+ More
A0A194RBG4 A0A212EJ41 A0A139WHM5 A0A139WHM0 E2BQP7 A0A0J7L2Y7 A0A1Y1LXV7 A0A1B6E7E5 A0A1B6BXW7 A0A1B6DYA5 A0A1B6E2D1 A0A0N0U7F7 A0A310SN20 A0A2R7VPZ1 A0A023EYJ1 A0A146L160 A0A146MGE9 T1HPP7 A0A146LWK8 A0A2S2R5X9 A0A336LJL6 A0A336LNX1
A0A194RBG4 A0A212EJ41 A0A139WHM5 A0A139WHM0 E2BQP7 A0A0J7L2Y7 A0A1Y1LXV7 A0A1B6E7E5 A0A1B6BXW7 A0A1B6DYA5 A0A1B6E2D1 A0A0N0U7F7 A0A310SN20 A0A2R7VPZ1 A0A023EYJ1 A0A146L160 A0A146MGE9 T1HPP7 A0A146LWK8 A0A2S2R5X9 A0A336LJL6 A0A336LNX1
Ontologies
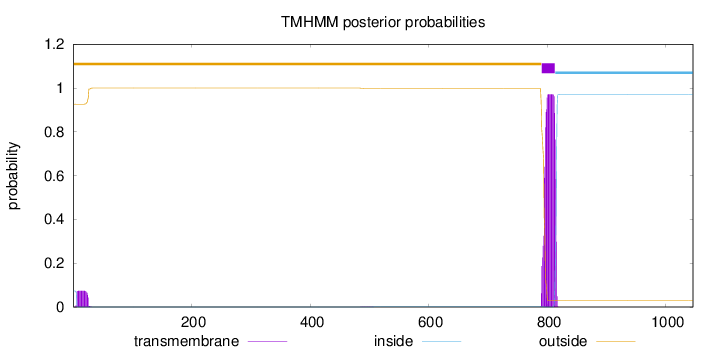
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.964614
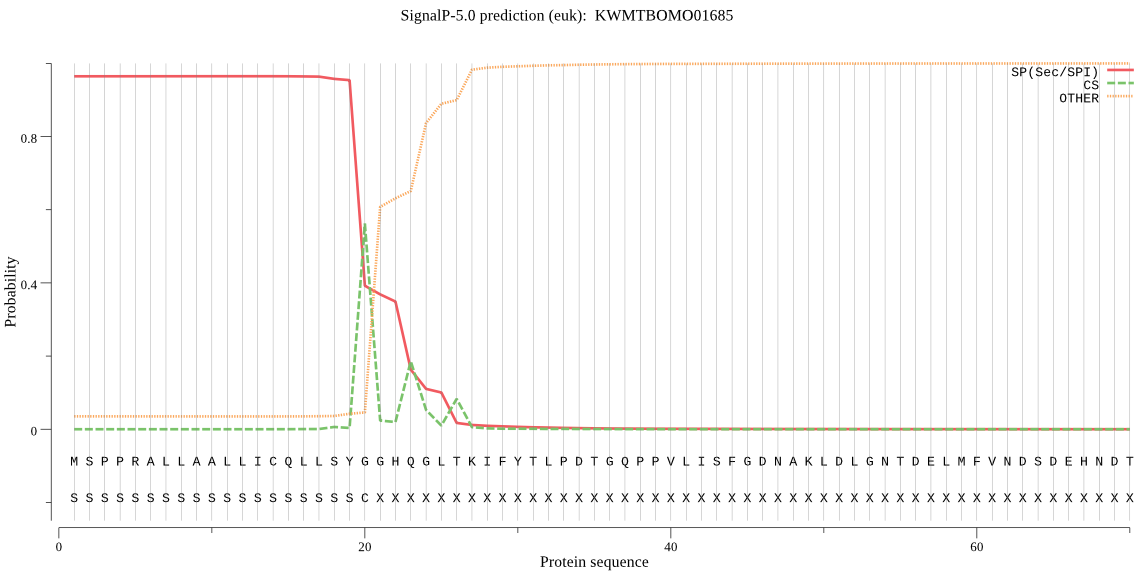
Length:
1046
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.558
Exp number, first 60 AAs:
1.42964
Total prob of N-in:
0.07368
outside
1 - 790
TMhelix
791 - 813
inside
814 - 1046
Population Genetic Test Statistics
Pi
209.52881
Theta
203.560592
Tajima's D
0.0599
CLR
0.597447
CSRT
0.380930953452327
Interpretation
Uncertain
