Gene
KWMTBOMO01683
Pre Gene Modal
BGIBMGA006037
Annotation
eIF_2a_kinase_[Bombyx_mori]
Full name
Eukaryotic translation initiation factor 2-alpha kinase 1
Alternative Name
Heme-regulated eukaryotic initiation factor eIF-2-alpha kinase
Hemin-sensitive initiation factor 2-alpha kinase
Heme-controlled repressor
Heme-regulated inhibitor
Hemin-sensitive initiation factor 2-alpha kinase
Heme-controlled repressor
Heme-regulated inhibitor
Location in the cell
Cytoplasmic Reliability : 2.941
Sequence
CDS
ATGGATAAACATAGCCAAGACAAATGGAAAGCATTGGCGACAGTGAAATCCTTCGATTTAGGCATATCGGCTAGTCACCATGAGTCATTCGTACAGCAGAGTAGACAACAGATTGATGTCATCAATGCCCCAACCACAACCCCAATCAGCCTCCTAGTTCAATCACTCGTTAAACAATTGTGTTCATTGTTACAAAAAGACAGTATTATAGCCAATCAGCTTTACAACAAAATATGTGAGAAACTTCATAGTATGAACTTGATTGACAATTCGTATGCTATGGGAGAGTTTGAAGCTATGAGAAGTCAATATCAGAGAGCCTTGTATCAGCTTGTGACGGTCGCCAGCGGAACAGAGATACCGATAATACTGCCAGCAACTTGGCCTATAGTTCAGCCGTCTGGACTTGAATGGTCAAGATACCATAGAGAATTTGAGGAGCTCTACTTCATAGCTGGTGGAGGCTTCGGGAGCGTTTTCAAAGCGCGACACAGACTAGATGGCGTAGAGTATGCCGTCAAAAAAGTTTACATTAAATCTTCAGACGTCGACTCTATCATGAGTCATTTGTCGGAAGTCAAAACAATAGCCAGTCTCAATCACCCGAACATAGTAAACTATAAGGCAGCGTGGCTCGAACCCATGATAGAGTCTACAGTTAAAAAGAAAGGCAAATATCAAATGGACACCGACAGTGACGAGTTTTCATTAAGCTCTGACCTCATATCATCAGCACATCCCAACGTAATAAATTCATTCAAGACTCACAACTCCAAGGAGTTGACCAAAAACAAGAGCCTGTCCGACTTCATTATTTCCTTCAAGAACTCCAACAGCTTCGAGAACTTGAACAGTTCGAACGAAGAACTGCAGGTTTCTGACAGCGATGACGAGTCCGTTTCGCAGGAGGAGAATGCCGTTTGCAATCTCTTTTCCAGTAAGGAGTACGAAAATTGCTCTCGTATAAACCTTAAATGGGCCACCTTGTTCATCCAGATGACGTTCTGCCAGCAAACCTTGAAGCAGTGGCTTGACGAGCGCAACAACCATATGTCAGTGTCGCGAAAAGGTTCCGACGATTTCACTCTGCCGTTACATGTGTGTGAATCTCCAATTGAGGCTAAAGATATCACGTTCCCAGCGAGTATTAATCACATTGACCTGCTGATAGACATGTTCACGCAGCTGGTGCGTGGTCTCCATTATATACATTCCCGTGGTATTATCCACCACGACATAAAGCCGAGTAATGTATTCGTCGCGCCACATGAAGGTGGCTTGTTGGTGCAGTTGGGTGATTTCGGTTTGGCTTGCCCGTTACAGCAGTCCCATAGTGGATTGGCACTCGGTACACATATGTATGCTGCACCGGAGCAACTGGATGGGCAGTGCAATCCAAAGAGCGACATGTACAGTCTGGGTATAATATTACTTGAGTTGGTAGAACCATTCGTGACTGATATGGAACGAGTGAAAACTATCACCGACCTCCGCAAAGGTCAGATTCCAGCTCACCTCACTGCCAACTACCCAAAAATTGCTCATATCATCGGCAAACTGGTGCAAAGGAAGCCCAGCAAGAGACTGGACACGGCCCAGCTGCTGGAGGAACTCAAGACCCTGGCCGAGAATAAAGATGACACGATCAGATCGTTGCGAGAGGAGCTCGCTGCGAAAGATGACGAAATAGCTAAACTCAAGATGATGCTGGCGAACTTGAATTTTAAATCTTCAGTGTGA
Protein
MDKHSQDKWKALATVKSFDLGISASHHESFVQQSRQQIDVINAPTTTPISLLVQSLVKQLCSLLQKDSIIANQLYNKICEKLHSMNLIDNSYAMGEFEAMRSQYQRALYQLVTVASGTEIPIILPATWPIVQPSGLEWSRYHREFEELYFIAGGGFGSVFKARHRLDGVEYAVKKVYIKSSDVDSIMSHLSEVKTIASLNHPNIVNYKAAWLEPMIESTVKKKGKYQMDTDSDEFSLSSDLISSAHPNVINSFKTHNSKELTKNKSLSDFIISFKNSNSFENLNSSNEELQVSDSDDESVSQEENAVCNLFSSKEYENCSRINLKWATLFIQMTFCQQTLKQWLDERNNHMSVSRKGSDDFTLPLHVCESPIEAKDITFPASINHIDLLIDMFTQLVRGLHYIHSRGIIHHDIKPSNVFVAPHEGGLLVQLGDFGLACPLQQSHSGLALGTHMYAAPEQLDGQCNPKSDMYSLGIILLELVEPFVTDMERVKTITDLRKGQIPAHLTANYPKIAHIIGKLVQRKPSKRLDTAQLLEELKTLAENKDDTIRSLREELAAKDDEIAKLKMMLANLNFKSSV
Summary
Description
Inhibits protein synthesis at the translation initiation level, in response to various stress conditions, including oxidative stress, heme deficiency, osmotic shock and heat shock. Exerts its function through the phosphorylation of EIF2S1 at 'Ser-48' and 'Ser-51', thus preventing its recycling. Binds hemin forming a 1:1 complex through a cysteine thiolate and histidine nitrogenous coordination. This binding occurs with moderate affinity, allowing it to sense the heme concentration within the cell. Thanks to this unique heme-sensing capacity, plays a crucial role to shut off protein synthesis during acute heme-deficient conditions. In red blood cells (RBCs), controls hemoglobin synthesis ensuring a coordinated regulation of the synthesis of its heme and globin moieties. Thus plays an essential protective role for RBC survival in anemias of iron deficiency. Similarly, in hepatocytes, involved in heme-mediated translational control of CYP2B and CYP3A and possibly other hepatic P450 cytochromes. May also contain ER stress during acute heme-deficient conditions (By similarity).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Synthesized in an inactive form that binds to the N-terminal domain of CDC37. Has to be associated with a multiprotein complex containing Hsp90, CDC37 and PPP5C for maturation and activation by autophosphorylation. The phosphatase PPP5C modulates this activation. Forms oligomers. Has been reported as a homodimer, as well as a hexamer in the absence of hemin. Converted to an inactive disulfide linked homodimer in the presence of hemin (By similarity).
Synthesized in an inactive form that binds to the N-terminal domain of CDC37. Has to be associated with a multiprotein complex containing Hsp90, CDC37 and PPP5C for maturation and activation by autophosphorylation. The phosphatase PPP5C modulates this activation. Forms oligomers. Has been reported as a non-covalently bound homodimer, as well as a hexamer in the absence of hemin. Converted to an inactive disulfide linked homodimer in the presence of hemin (By similarity).
Synthesized in an inactive form that binds to the N-terminal domain of CDC37. Has to be associated with a multiprotein complex containing Hsp90, CDC37 and PPP5C for maturation and activation by autophosphorylation. The phosphatase PPP5C modulates this activation. Forms oligomers. Has been reported as a non-covalently bound homodimer, as well as a hexamer in the absence of hemin. Converted to an inactive disulfide linked homodimer in the presence of hemin (By similarity).
Miscellaneous
Can bind 1 molecules of heme per polypeptide chain.
Similarity
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. GCN2 subfamily.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Disulfide bond
Kinase
Nucleotide-binding
Phosphoprotein
Protein synthesis inhibitor
Repeat
Transferase
Alternative splicing
Polymorphism
Reference proteome
Feature
chain Eukaryotic translation initiation factor 2-alpha kinase 1
splice variant In isoform 2.
sequence variant In dbSNP:rs34889754.
splice variant In isoform 2.
sequence variant In dbSNP:rs34889754.
Uniprot
Q8I739
H9J945
A0A2H1VIE0
A0A2W1BNF1
A0A2A4KAZ2
A0A194R5Z6
+ More
A0A212EJ31 A0A194PYU9 A0A0L7L9L3 A0A336K8N1 A0A182HCR7 A0A182LSK0 A0A1W4WZY9 Q17MW3 D6WJR1 A0A084WAM0 A0A1B0GM98 A0A0K8TMR2 A0A146LBU4 A0A0A9WWL4 A0A1Y1KBL7 A0A026WQ75 A0A3L8DNL7 A0A158NV55 A0A195DZQ1 A0A1B6EJ28 A0A1B6I9A0 A0A1B6LV80 A0A096N681 F7A5Z1 G7P286 F7F136 A0A2K5MH01 Q4R8E0 H9EYQ6 A0A2K5MGV9 A0A2K5YAW5 A0A2K5X5T1 A0A2K5MGW2 Q9BQI3-2 A0A024QZU1 Q9BQI3 A0A2K6NPN8 A0A2R7X2V0 S7NHA6 A0A340WPE5 A0A0D9RYR1 A0A2K5SJB8 A0A2K6TJ62 A0A2R9BQ21 B4DIP4 H2QU55 K7DI71 A0A1U7UYU5 A0A2I3SA78 A0A1U7ULN9 A0A3Q0DDA5
A0A212EJ31 A0A194PYU9 A0A0L7L9L3 A0A336K8N1 A0A182HCR7 A0A182LSK0 A0A1W4WZY9 Q17MW3 D6WJR1 A0A084WAM0 A0A1B0GM98 A0A0K8TMR2 A0A146LBU4 A0A0A9WWL4 A0A1Y1KBL7 A0A026WQ75 A0A3L8DNL7 A0A158NV55 A0A195DZQ1 A0A1B6EJ28 A0A1B6I9A0 A0A1B6LV80 A0A096N681 F7A5Z1 G7P286 F7F136 A0A2K5MH01 Q4R8E0 H9EYQ6 A0A2K5MGV9 A0A2K5YAW5 A0A2K5X5T1 A0A2K5MGW2 Q9BQI3-2 A0A024QZU1 Q9BQI3 A0A2K6NPN8 A0A2R7X2V0 S7NHA6 A0A340WPE5 A0A0D9RYR1 A0A2K5SJB8 A0A2K6TJ62 A0A2R9BQ21 B4DIP4 H2QU55 K7DI71 A0A1U7UYU5 A0A2I3SA78 A0A1U7ULN9 A0A3Q0DDA5
EC Number
2.7.11.1
Pubmed
19121390
28756777
26354079
22118469
26227816
26483478
+ More
17510324 18362917 19820115 24438588 26369729 26823975 25401762 28004739 24508170 30249741 21347285 22002653 17431167 25319552 11101152 10718198 10931946 11230166 12391722 14702039 17974005 12690205 15489334 11036079 18691976 20071449 21269460 17344846 11181995 25362486 22722832 16136131
17510324 18362917 19820115 24438588 26369729 26823975 25401762 28004739 24508170 30249741 21347285 22002653 17431167 25319552 11101152 10718198 10931946 11230166 12391722 14702039 17974005 12690205 15489334 11036079 18691976 20071449 21269460 17344846 11181995 25362486 22722832 16136131
EMBL
U87236
AAO13686.1
BABH01004453
ODYU01002731
SOQ40595.1
KZ150052
+ More
PZC74396.1 NWSH01000011 PCG80910.1 KQ460644 KPJ13243.1 AGBW02014536 OWR41489.1 KQ459584 KPI98512.1 JTDY01002109 KOB72085.1 UFQS01000117 UFQT01000117 SSW99988.1 SSX20368.1 JXUM01034487 KQ561024 KXJ79985.1 AXCM01005219 CH477203 EAT48039.1 KQ971343 EFA03110.1 ATLV01022209 KE525330 KFB47264.1 AJVK01011228 GDAI01001974 JAI15629.1 GDHC01012796 JAQ05833.1 GBHO01032691 JAG10913.1 GEZM01091386 JAV56866.1 KK107135 EZA58098.1 QOIP01000006 RLU22024.1 ADTU01026895 KQ980031 KYN18182.1 GECZ01031845 JAS37924.1 GECU01024195 JAS83511.1 GEBQ01031866 GEBQ01012422 GEBQ01007005 JAT08111.1 JAT27555.1 JAT32972.1 CM001278 EHH51994.1 JSUE03027721 JSUE03027722 JSUE03027723 AB168513 JU323759 AFE67515.1 AQIA01045269 AQIA01045270 AF255050 AF100784 AF181071 AB037790 AF183414 AL136563 AF147094 AK075192 AK290327 AL834494 CH236963 CH878731 BC006524 AF116634 EAW55051.1 KK856629 PTY26137.1 KE164198 EPQ15910.1 AQIB01149654 AQIB01149655 AQIB01149656 AJFE02118599 AJFE02118600 AJFE02118601 AJFE02118602 AJFE02118603 AJFE02118604 AJFE02118605 AK295712 BAG58556.1 AC184156 GABC01003448 GABC01003447 NBAG03000572 JAA07890.1 PNI14766.1 GABF01003862 GABF01003861 GABD01000763 GABD01000762 GABE01005524 JAA18283.1 JAA32337.1 JAA39215.1
PZC74396.1 NWSH01000011 PCG80910.1 KQ460644 KPJ13243.1 AGBW02014536 OWR41489.1 KQ459584 KPI98512.1 JTDY01002109 KOB72085.1 UFQS01000117 UFQT01000117 SSW99988.1 SSX20368.1 JXUM01034487 KQ561024 KXJ79985.1 AXCM01005219 CH477203 EAT48039.1 KQ971343 EFA03110.1 ATLV01022209 KE525330 KFB47264.1 AJVK01011228 GDAI01001974 JAI15629.1 GDHC01012796 JAQ05833.1 GBHO01032691 JAG10913.1 GEZM01091386 JAV56866.1 KK107135 EZA58098.1 QOIP01000006 RLU22024.1 ADTU01026895 KQ980031 KYN18182.1 GECZ01031845 JAS37924.1 GECU01024195 JAS83511.1 GEBQ01031866 GEBQ01012422 GEBQ01007005 JAT08111.1 JAT27555.1 JAT32972.1 CM001278 EHH51994.1 JSUE03027721 JSUE03027722 JSUE03027723 AB168513 JU323759 AFE67515.1 AQIA01045269 AQIA01045270 AF255050 AF100784 AF181071 AB037790 AF183414 AL136563 AF147094 AK075192 AK290327 AL834494 CH236963 CH878731 BC006524 AF116634 EAW55051.1 KK856629 PTY26137.1 KE164198 EPQ15910.1 AQIB01149654 AQIB01149655 AQIB01149656 AJFE02118599 AJFE02118600 AJFE02118601 AJFE02118602 AJFE02118603 AJFE02118604 AJFE02118605 AK295712 BAG58556.1 AC184156 GABC01003448 GABC01003447 NBAG03000572 JAA07890.1 PNI14766.1 GABF01003862 GABF01003861 GABD01000763 GABD01000762 GABE01005524 JAA18283.1 JAA32337.1 JAA39215.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000069940 UP000249989 UP000075883 UP000192223 UP000008820 UP000007266 UP000030765 UP000092462 UP000053097 UP000279307 UP000005205 UP000078492 UP000028761 UP000008225 UP000009130 UP000006718 UP000233060 UP000233100 UP000233140 UP000005640 UP000233200 UP000265300 UP000029965 UP000233040 UP000233220 UP000240080 UP000002277 UP000189704
UP000069940 UP000249989 UP000075883 UP000192223 UP000008820 UP000007266 UP000030765 UP000092462 UP000053097 UP000279307 UP000005205 UP000078492 UP000028761 UP000008225 UP000009130 UP000006718 UP000233060 UP000233100 UP000233140 UP000005640 UP000233200 UP000265300 UP000029965 UP000233040 UP000233220 UP000240080 UP000002277 UP000189704
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Q8I739
H9J945
A0A2H1VIE0
A0A2W1BNF1
A0A2A4KAZ2
A0A194R5Z6
+ More
A0A212EJ31 A0A194PYU9 A0A0L7L9L3 A0A336K8N1 A0A182HCR7 A0A182LSK0 A0A1W4WZY9 Q17MW3 D6WJR1 A0A084WAM0 A0A1B0GM98 A0A0K8TMR2 A0A146LBU4 A0A0A9WWL4 A0A1Y1KBL7 A0A026WQ75 A0A3L8DNL7 A0A158NV55 A0A195DZQ1 A0A1B6EJ28 A0A1B6I9A0 A0A1B6LV80 A0A096N681 F7A5Z1 G7P286 F7F136 A0A2K5MH01 Q4R8E0 H9EYQ6 A0A2K5MGV9 A0A2K5YAW5 A0A2K5X5T1 A0A2K5MGW2 Q9BQI3-2 A0A024QZU1 Q9BQI3 A0A2K6NPN8 A0A2R7X2V0 S7NHA6 A0A340WPE5 A0A0D9RYR1 A0A2K5SJB8 A0A2K6TJ62 A0A2R9BQ21 B4DIP4 H2QU55 K7DI71 A0A1U7UYU5 A0A2I3SA78 A0A1U7ULN9 A0A3Q0DDA5
A0A212EJ31 A0A194PYU9 A0A0L7L9L3 A0A336K8N1 A0A182HCR7 A0A182LSK0 A0A1W4WZY9 Q17MW3 D6WJR1 A0A084WAM0 A0A1B0GM98 A0A0K8TMR2 A0A146LBU4 A0A0A9WWL4 A0A1Y1KBL7 A0A026WQ75 A0A3L8DNL7 A0A158NV55 A0A195DZQ1 A0A1B6EJ28 A0A1B6I9A0 A0A1B6LV80 A0A096N681 F7A5Z1 G7P286 F7F136 A0A2K5MH01 Q4R8E0 H9EYQ6 A0A2K5MGV9 A0A2K5YAW5 A0A2K5X5T1 A0A2K5MGW2 Q9BQI3-2 A0A024QZU1 Q9BQI3 A0A2K6NPN8 A0A2R7X2V0 S7NHA6 A0A340WPE5 A0A0D9RYR1 A0A2K5SJB8 A0A2K6TJ62 A0A2R9BQ21 B4DIP4 H2QU55 K7DI71 A0A1U7UYU5 A0A2I3SA78 A0A1U7ULN9 A0A3Q0DDA5
PDB
4M7I
E-value=4.29382e-23,
Score=269
Ontologies
GO
GO:0004672
GO:0005524
GO:0003743
GO:0004694
GO:1990641
GO:0002526
GO:0006909
GO:0008285
GO:0046501
GO:0042803
GO:0005737
GO:0010999
GO:0030225
GO:0055072
GO:0020037
GO:0046986
GO:0046777
GO:0004674
GO:0017148
GO:0045993
GO:0016301
GO:0006468
GO:0006355
GO:0006508
GO:0006811
GO:0016020
GO:0005515
GO:0003779
GO:0016021
Topology
Subcellular location
Cytoplasm
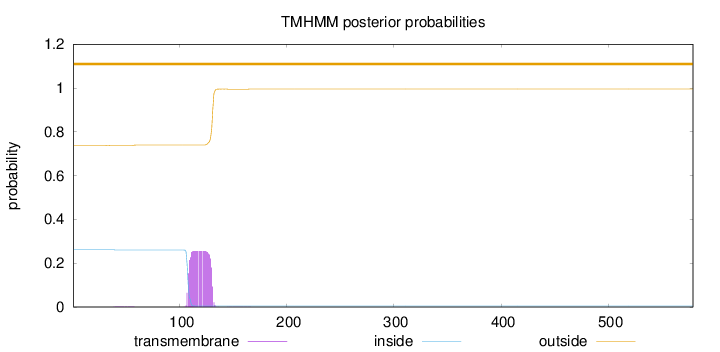
Length:
579
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.79285999999998
Exp number, first 60 AAs:
0.02606
Total prob of N-in:
0.26164
outside
1 - 579
Population Genetic Test Statistics
Pi
156.127866
Theta
182.287761
Tajima's D
-0.709259
CLR
0.595834
CSRT
0.189890505474726
Interpretation
Uncertain
