Gene
KWMTBOMO01681
Pre Gene Modal
BGIBMGA006111
Annotation
PREDICTED:_dual_specificity_tyrosine-phosphorylation-regulated_kinase_2-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.64
Sequence
CDS
ATGGAGGTACGTATAAAAGGAATTTACGAAATTGAAGCACTGAAGCTGTACGGCGAAAGACTAACGGAATGGGAGAGGAACGAGATAAGGAAGTACCCTCAGATCTGGTTCCTTGGCTTGGAGTCCAACAAGGTACAGGCTAGGTCCAACCTGCCTAACAATTGCGGGTTCGACGATGAAAACGGGAGTTATAACAAGCAAATCCACGACCACATCGCGTACAGATACGAGATCCTCGAAATAATAGGGAAAGGTTCGTTCGGGCAAGTTATCAGAGCTCTCGACCATCGCACCGGTGAGCAGGTAGCGATCAAAATTATCAGGAACAAGAAGCGCTTCCACCACCAGGCCCTGGTTGAAGTCAGGGTGTTGGATCACCTCCGATTGAAAGACAAGGACCAGTCGCATAACGTGATCCACATGCTGGATTACTTTTACTTCAGGAATCATTTATGCATTAGTTTCGAATTAATGAGCATCAACTTGTACGAGCTGATAAAGAAGAATAACTATCAGGGCTTCAGTTTGAGCCTCATCAGAAGGTTCGCTAGCTCGTTACTGCGCTGTCTTCGGCTCCTCGAAGCTGAGAACATCATACACTGCGACTTGAAACCGGAGAACATCTTGCTGAAGGCCAGGGGCAGTTCCTCCATCAAGGTGATTGACTTCGGCAGCTCCTGTTACACAGACGCTCGTGTCTACACATACATACAGTCGCGATTCTACCGGAGTCCCGAAGTCATACTAGGGTTGCAGTACGGACCGGCCATTGATATGTGGAGTATGGGGTGTATCCTTGCGGAACTTCACACGGGCTATCCCTTGTTCCCGGGAGAGAACGAGGCCGAACAACTAGCCTGCATCATGGAGCTGCTCGGACCGCCTCCACCAGACCTGCTAATCCACGCTACCAGAAAGAGGCTTTTCTTCGATTCGAAAGGTTCTCCTCGAAGCGTGACGAACTCAAAAGGAAGAAAACGCAGACCCGGGTCAAGGACTTTAGCGTCGGCCGTCAGAACCGAAGAGCCGGCCTTCCTGCACTTCCTCCACCGGTGCTTAGAATGGGATCCGAAGCGTCGTATAACACCAACGGAGGCGTCCCGCCACGAGTTCGTGACCGGCTGTCCTCTGGGAAGTACACCCTCGGGAGTCCTCGCGTTACCACGTCTCGCCCCGCCACGGCCAACACGCGGGGCGCTACTGCACTCTCAGTCTGCCGGCGACGTGGCTGCGATGCTCGGCCGCGCGTGA
Protein
MEVRIKGIYEIEALKLYGERLTEWERNEIRKYPQIWFLGLESNKVQARSNLPNNCGFDDENGSYNKQIHDHIAYRYEILEIIGKGSFGQVIRALDHRTGEQVAIKIIRNKKRFHHQALVEVRVLDHLRLKDKDQSHNVIHMLDYFYFRNHLCISFELMSINLYELIKKNNYQGFSLSLIRRFASSLLRCLRLLEAENIIHCDLKPENILLKARGSSSIKVIDFGSSCYTDARVYTYIQSRFYRSPEVILGLQYGPAIDMWSMGCILAELHTGYPLFPGENEAEQLACIMELLGPPPPDLLIHATRKRLFFDSKGSPRSVTNSKGRKRRPGSRTLASAVRTEEPAFLHFLHRCLEWDPKRRITPTEASRHEFVTGCPLGSTPSGVLALPRLAPPRPTRGALLHSQSAGDVAAMLGRA
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A212EJ25
A0A2W1BH26
A0A2A4KAI1
A0A194Q522
A0A194R7N1
H9J9B9
+ More
A0A1Y1LEW8 A0A0P5H690 A0A0P6HRP8 A0A0P5DKI9 A0A0P6AXI5 A0A0P5DER7 A0A1B0D2T7 A0A0P5PBK0 A0A1L8DKI9 A0A1L8DKJ8 A0A0T6AYV9 T1IV67 A0A2P8YEX4 K7IWI5 U5EXE3 A0A0A9ZJS4 Q17L44 A0A182G7L4 A0A1B6IP59 A0A151WKP4 A0A146KNU9 A0A1B6HM39 A0A0A9W1C5 A0A158NXL8 A0A151IPB2 A0A151I2M2 A0A195ECK3 F4WQ81 A0A2J7PUQ0 E2BZA9 A0A1B6DNE8 A0A2J7PUP5 A0A1B6DY09 E2AXX9 A0A026WM24 A0A3L8E2F3 E9GL45 A0A1B6M0U2 A0A224XMM8 A0A1B6M9U2 A0A069DVQ7 A0A0P4VM65 A0A023F3C9 A0A195EUJ4 A0A2R7WS36 A0A067RE91 A0A0C9R928 A0A0C9RN31 A0A182F606 A0A2A3EG10 A0A154PEA2 A0A2M4CJR2 E0VVI8 A0A1S4GWF8 A0A084W3W2 A0A1S4GWE7 A0A182W9Y1 A0A182IUA7 A0A182WTH2 A0A182P0I3 A0A182R3A5 A0A182IEL6 A0A182V7Y2 A0A182NLL8 A0A1W5BGI3 A0A182L2C8 A0A139WI49 D6WKF6 A0A139WIC6 A0A0P4ZMA0 A0A1W2W8T0 A0A147BM13 A0A1B0CJL0 C3Y4L0 A0A1B6GLS3 H2YKY2 A0A1B6EXG0 T1HR78 A0A2B4SBZ4 A0A2H1WTK6 W5NIX2 W5NIX4 V4APG6 A0A2R8QL82 A0A0L0BZ51 A0A060W305 A0A3P8YBL5 A0A2R8PY13 A0A2R8RWZ6 A0A3P8YA21 A0A3P8YA73 E7F1S5 A0A3B3DX97 A0A0R4IT26
A0A1Y1LEW8 A0A0P5H690 A0A0P6HRP8 A0A0P5DKI9 A0A0P6AXI5 A0A0P5DER7 A0A1B0D2T7 A0A0P5PBK0 A0A1L8DKI9 A0A1L8DKJ8 A0A0T6AYV9 T1IV67 A0A2P8YEX4 K7IWI5 U5EXE3 A0A0A9ZJS4 Q17L44 A0A182G7L4 A0A1B6IP59 A0A151WKP4 A0A146KNU9 A0A1B6HM39 A0A0A9W1C5 A0A158NXL8 A0A151IPB2 A0A151I2M2 A0A195ECK3 F4WQ81 A0A2J7PUQ0 E2BZA9 A0A1B6DNE8 A0A2J7PUP5 A0A1B6DY09 E2AXX9 A0A026WM24 A0A3L8E2F3 E9GL45 A0A1B6M0U2 A0A224XMM8 A0A1B6M9U2 A0A069DVQ7 A0A0P4VM65 A0A023F3C9 A0A195EUJ4 A0A2R7WS36 A0A067RE91 A0A0C9R928 A0A0C9RN31 A0A182F606 A0A2A3EG10 A0A154PEA2 A0A2M4CJR2 E0VVI8 A0A1S4GWF8 A0A084W3W2 A0A1S4GWE7 A0A182W9Y1 A0A182IUA7 A0A182WTH2 A0A182P0I3 A0A182R3A5 A0A182IEL6 A0A182V7Y2 A0A182NLL8 A0A1W5BGI3 A0A182L2C8 A0A139WI49 D6WKF6 A0A139WIC6 A0A0P4ZMA0 A0A1W2W8T0 A0A147BM13 A0A1B0CJL0 C3Y4L0 A0A1B6GLS3 H2YKY2 A0A1B6EXG0 T1HR78 A0A2B4SBZ4 A0A2H1WTK6 W5NIX2 W5NIX4 V4APG6 A0A2R8QL82 A0A0L0BZ51 A0A060W305 A0A3P8YBL5 A0A2R8PY13 A0A2R8RWZ6 A0A3P8YA21 A0A3P8YA73 E7F1S5 A0A3B3DX97 A0A0R4IT26
Pubmed
22118469
28756777
26354079
19121390
28004739
29403074
+ More
20075255 17510324 26483478 26823975 25401762 21347285 21719571 20798317 24508170 30249741 21292972 26334808 27129103 25474469 24845553 20566863 12364791 24438588 20966253 18362917 19820115 29652888 18563158 23254933 23594743 26108605 24755649 25069045 29451363
20075255 17510324 26483478 26823975 25401762 21347285 21719571 20798317 24508170 30249741 21292972 26334808 27129103 25474469 24845553 20566863 12364791 24438588 20966253 18362917 19820115 29652888 18563158 23254933 23594743 26108605 24755649 25069045 29451363
EMBL
AGBW02014536
OWR41487.1
KZ150052
PZC74399.1
NWSH01000011
PCG80908.1
+ More
KQ459584 KPI98510.1 KQ460644 KPJ13245.1 BABH01004443 BABH01004444 BABH01004445 BABH01004446 GEZM01057673 JAV72164.1 GDIQ01238375 JAK13350.1 GDIQ01014346 JAN80391.1 GDIP01171863 JAJ51539.1 GDIP01022908 JAM80807.1 GDIP01157266 JAJ66136.1 AJVK01002814 AJVK01002815 GDIQ01133108 JAL18618.1 GFDF01007105 JAV06979.1 GFDF01007096 JAV06988.1 LJIG01022485 KRT80329.1 JH431575 PYGN01000651 PSN42817.1 GANO01000990 JAB58881.1 GBRD01003605 JAG62216.1 CH477219 EAT47382.1 JXUM01047004 JXUM01047005 JXUM01047006 JXUM01047007 JXUM01047008 JXUM01047009 JXUM01047010 JXUM01047011 JXUM01047012 JXUM01047013 GECU01018988 JAS88718.1 KQ983001 KYQ48428.1 GDHC01020880 JAP97748.1 GECU01031957 JAS75749.1 GBHO01043336 JAG00268.1 ADTU01003149 ADTU01003150 ADTU01003151 ADTU01003152 ADTU01003153 ADTU01003154 ADTU01003155 ADTU01003156 ADTU01003157 ADTU01003158 KQ976877 KYN07677.1 KQ976520 KYM82095.1 KQ979074 KYN22943.1 GL888264 EGI63627.1 NEVH01021193 PNF20059.1 GL451577 EFN79015.1 GEDC01010109 JAS27189.1 PNF20057.1 GEDC01006779 JAS30519.1 GL443740 EFN61682.1 KK107168 EZA56139.1 QOIP01000001 RLU26867.1 GL732550 EFX79858.1 GEBQ01010442 JAT29535.1 GFTR01007033 JAW09393.1 GEBQ01007275 JAT32702.1 GBGD01001102 JAC87787.1 GDKW01000981 JAI55614.1 GBBI01002986 JAC15726.1 KQ981958 KYN31925.1 KK855397 PTY22366.1 KK852691 KDR18363.1 GBYB01004660 JAG74427.1 GBYB01008481 JAG78248.1 KZ288267 PBC30086.1 KQ434886 KZC10173.1 GGFL01001389 MBW65567.1 DS235812 EEB17394.1 AAAB01008964 ATLV01020170 ATLV01020171 ATLV01020172 KE525295 KFB44906.1 APCN01005209 KQ971342 KYB27653.1 EFA03986.2 KYB27654.1 GDIP01211311 JAJ12091.1 GEGO01003892 JAR91512.1 AJWK01014701 GG666486 EEN64667.1 GECZ01006384 JAS63385.1 GECZ01027100 JAS42669.1 ACPB03024912 LSMT01000134 PFX26108.1 ODYU01010959 SOQ56391.1 AHAT01025779 AHAT01025780 AHAT01025781 KB201656 ESO95526.1 CR396584 JRES01001125 KNC25276.1 FR904385 CDQ61648.1
KQ459584 KPI98510.1 KQ460644 KPJ13245.1 BABH01004443 BABH01004444 BABH01004445 BABH01004446 GEZM01057673 JAV72164.1 GDIQ01238375 JAK13350.1 GDIQ01014346 JAN80391.1 GDIP01171863 JAJ51539.1 GDIP01022908 JAM80807.1 GDIP01157266 JAJ66136.1 AJVK01002814 AJVK01002815 GDIQ01133108 JAL18618.1 GFDF01007105 JAV06979.1 GFDF01007096 JAV06988.1 LJIG01022485 KRT80329.1 JH431575 PYGN01000651 PSN42817.1 GANO01000990 JAB58881.1 GBRD01003605 JAG62216.1 CH477219 EAT47382.1 JXUM01047004 JXUM01047005 JXUM01047006 JXUM01047007 JXUM01047008 JXUM01047009 JXUM01047010 JXUM01047011 JXUM01047012 JXUM01047013 GECU01018988 JAS88718.1 KQ983001 KYQ48428.1 GDHC01020880 JAP97748.1 GECU01031957 JAS75749.1 GBHO01043336 JAG00268.1 ADTU01003149 ADTU01003150 ADTU01003151 ADTU01003152 ADTU01003153 ADTU01003154 ADTU01003155 ADTU01003156 ADTU01003157 ADTU01003158 KQ976877 KYN07677.1 KQ976520 KYM82095.1 KQ979074 KYN22943.1 GL888264 EGI63627.1 NEVH01021193 PNF20059.1 GL451577 EFN79015.1 GEDC01010109 JAS27189.1 PNF20057.1 GEDC01006779 JAS30519.1 GL443740 EFN61682.1 KK107168 EZA56139.1 QOIP01000001 RLU26867.1 GL732550 EFX79858.1 GEBQ01010442 JAT29535.1 GFTR01007033 JAW09393.1 GEBQ01007275 JAT32702.1 GBGD01001102 JAC87787.1 GDKW01000981 JAI55614.1 GBBI01002986 JAC15726.1 KQ981958 KYN31925.1 KK855397 PTY22366.1 KK852691 KDR18363.1 GBYB01004660 JAG74427.1 GBYB01008481 JAG78248.1 KZ288267 PBC30086.1 KQ434886 KZC10173.1 GGFL01001389 MBW65567.1 DS235812 EEB17394.1 AAAB01008964 ATLV01020170 ATLV01020171 ATLV01020172 KE525295 KFB44906.1 APCN01005209 KQ971342 KYB27653.1 EFA03986.2 KYB27654.1 GDIP01211311 JAJ12091.1 GEGO01003892 JAR91512.1 AJWK01014701 GG666486 EEN64667.1 GECZ01006384 JAS63385.1 GECZ01027100 JAS42669.1 ACPB03024912 LSMT01000134 PFX26108.1 ODYU01010959 SOQ56391.1 AHAT01025779 AHAT01025780 AHAT01025781 KB201656 ESO95526.1 CR396584 JRES01001125 KNC25276.1 FR904385 CDQ61648.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000005204
UP000092462
+ More
UP000245037 UP000002358 UP000008820 UP000069940 UP000075809 UP000005205 UP000078542 UP000078540 UP000078492 UP000007755 UP000235965 UP000008237 UP000000311 UP000053097 UP000279307 UP000000305 UP000078541 UP000027135 UP000069272 UP000242457 UP000076502 UP000009046 UP000030765 UP000075920 UP000075880 UP000076407 UP000075885 UP000075900 UP000075840 UP000075903 UP000075884 UP000075882 UP000007266 UP000092461 UP000001554 UP000007875 UP000015103 UP000225706 UP000018468 UP000030746 UP000000437 UP000037069 UP000193380 UP000265140 UP000261560
UP000245037 UP000002358 UP000008820 UP000069940 UP000075809 UP000005205 UP000078542 UP000078540 UP000078492 UP000007755 UP000235965 UP000008237 UP000000311 UP000053097 UP000279307 UP000000305 UP000078541 UP000027135 UP000069272 UP000242457 UP000076502 UP000009046 UP000030765 UP000075920 UP000075880 UP000076407 UP000075885 UP000075900 UP000075840 UP000075903 UP000075884 UP000075882 UP000007266 UP000092461 UP000001554 UP000007875 UP000015103 UP000225706 UP000018468 UP000030746 UP000000437 UP000037069 UP000193380 UP000265140 UP000261560
PRIDE
Pfam
Interpro
IPR017441
Protein_kinase_ATP_BS
+ More
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR001478 PDZ
IPR036034 PDZ_sf
IPR003961 FN3_dom
IPR011333 SKP1/BTB/POZ_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR000210 BTB/POZ_dom
IPR013783 Ig-like_fold
IPR006652 Kelch_1
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR015915 Kelch-typ_b-propeller
IPR003598 Ig_sub2
IPR011705 BACK
IPR036179 Ig-like_dom_sf
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR001478 PDZ
IPR036034 PDZ_sf
IPR003961 FN3_dom
IPR011333 SKP1/BTB/POZ_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR000210 BTB/POZ_dom
IPR013783 Ig-like_fold
IPR006652 Kelch_1
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR015915 Kelch-typ_b-propeller
IPR003598 Ig_sub2
IPR011705 BACK
IPR036179 Ig-like_dom_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A212EJ25
A0A2W1BH26
A0A2A4KAI1
A0A194Q522
A0A194R7N1
H9J9B9
+ More
A0A1Y1LEW8 A0A0P5H690 A0A0P6HRP8 A0A0P5DKI9 A0A0P6AXI5 A0A0P5DER7 A0A1B0D2T7 A0A0P5PBK0 A0A1L8DKI9 A0A1L8DKJ8 A0A0T6AYV9 T1IV67 A0A2P8YEX4 K7IWI5 U5EXE3 A0A0A9ZJS4 Q17L44 A0A182G7L4 A0A1B6IP59 A0A151WKP4 A0A146KNU9 A0A1B6HM39 A0A0A9W1C5 A0A158NXL8 A0A151IPB2 A0A151I2M2 A0A195ECK3 F4WQ81 A0A2J7PUQ0 E2BZA9 A0A1B6DNE8 A0A2J7PUP5 A0A1B6DY09 E2AXX9 A0A026WM24 A0A3L8E2F3 E9GL45 A0A1B6M0U2 A0A224XMM8 A0A1B6M9U2 A0A069DVQ7 A0A0P4VM65 A0A023F3C9 A0A195EUJ4 A0A2R7WS36 A0A067RE91 A0A0C9R928 A0A0C9RN31 A0A182F606 A0A2A3EG10 A0A154PEA2 A0A2M4CJR2 E0VVI8 A0A1S4GWF8 A0A084W3W2 A0A1S4GWE7 A0A182W9Y1 A0A182IUA7 A0A182WTH2 A0A182P0I3 A0A182R3A5 A0A182IEL6 A0A182V7Y2 A0A182NLL8 A0A1W5BGI3 A0A182L2C8 A0A139WI49 D6WKF6 A0A139WIC6 A0A0P4ZMA0 A0A1W2W8T0 A0A147BM13 A0A1B0CJL0 C3Y4L0 A0A1B6GLS3 H2YKY2 A0A1B6EXG0 T1HR78 A0A2B4SBZ4 A0A2H1WTK6 W5NIX2 W5NIX4 V4APG6 A0A2R8QL82 A0A0L0BZ51 A0A060W305 A0A3P8YBL5 A0A2R8PY13 A0A2R8RWZ6 A0A3P8YA21 A0A3P8YA73 E7F1S5 A0A3B3DX97 A0A0R4IT26
A0A1Y1LEW8 A0A0P5H690 A0A0P6HRP8 A0A0P5DKI9 A0A0P6AXI5 A0A0P5DER7 A0A1B0D2T7 A0A0P5PBK0 A0A1L8DKI9 A0A1L8DKJ8 A0A0T6AYV9 T1IV67 A0A2P8YEX4 K7IWI5 U5EXE3 A0A0A9ZJS4 Q17L44 A0A182G7L4 A0A1B6IP59 A0A151WKP4 A0A146KNU9 A0A1B6HM39 A0A0A9W1C5 A0A158NXL8 A0A151IPB2 A0A151I2M2 A0A195ECK3 F4WQ81 A0A2J7PUQ0 E2BZA9 A0A1B6DNE8 A0A2J7PUP5 A0A1B6DY09 E2AXX9 A0A026WM24 A0A3L8E2F3 E9GL45 A0A1B6M0U2 A0A224XMM8 A0A1B6M9U2 A0A069DVQ7 A0A0P4VM65 A0A023F3C9 A0A195EUJ4 A0A2R7WS36 A0A067RE91 A0A0C9R928 A0A0C9RN31 A0A182F606 A0A2A3EG10 A0A154PEA2 A0A2M4CJR2 E0VVI8 A0A1S4GWF8 A0A084W3W2 A0A1S4GWE7 A0A182W9Y1 A0A182IUA7 A0A182WTH2 A0A182P0I3 A0A182R3A5 A0A182IEL6 A0A182V7Y2 A0A182NLL8 A0A1W5BGI3 A0A182L2C8 A0A139WI49 D6WKF6 A0A139WIC6 A0A0P4ZMA0 A0A1W2W8T0 A0A147BM13 A0A1B0CJL0 C3Y4L0 A0A1B6GLS3 H2YKY2 A0A1B6EXG0 T1HR78 A0A2B4SBZ4 A0A2H1WTK6 W5NIX2 W5NIX4 V4APG6 A0A2R8QL82 A0A0L0BZ51 A0A060W305 A0A3P8YBL5 A0A2R8PY13 A0A2R8RWZ6 A0A3P8YA21 A0A3P8YA73 E7F1S5 A0A3B3DX97 A0A0R4IT26
Ontologies
GO
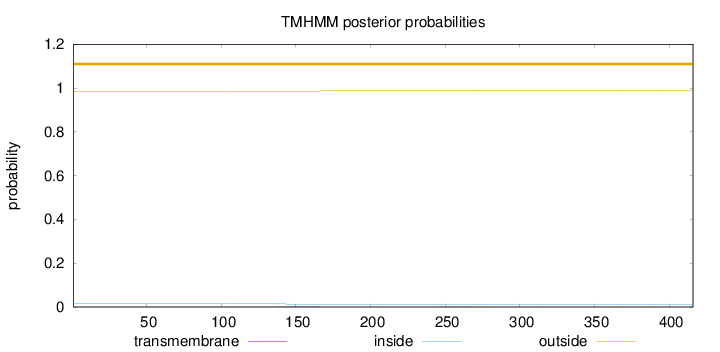
Topology
Length:
416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07887
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01468
outside
1 - 416
Population Genetic Test Statistics
Pi
176.363636
Theta
153.145816
Tajima's D
0.274909
CLR
0.401572
CSRT
0.449177541122944
Interpretation
Uncertain
