Gene
KWMTBOMO01677
Pre Gene Modal
BGIBMGA006038
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_7_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.848 Nuclear Reliability : 2.17
Sequence
CDS
ATGGATGGATCCTGCAAGATCTTGCACGAAGAACTGGCGCTGCAGTGGGTGGTCGCCAGTGGAGTGACCAGGGACCTCGCCATGCAGAACTCCTGGGCGCTGTTCGAGCTGCTGGTGAAGTCGATGTCGGAGTACGTGCAGTGGAGCGGCGCGCACGAGGCGGCCCGCAAGGCGCGCTTCCCGGAGCCCTTCACCGACGACCTCACCACCCTCGTCAACTGCGTCACCTCCGAGATCATATCCGGCTACGGCAAGAACAGTCGGCTGACGCAGAGCCTCAACAACAGCCTGGCGTTCTTCCTGTTCGACCTCATCAACATAATGGACCGCGGCTACGTGCTGAGCCTCGTCCGGACCTACTACAAGCAGATGAGCGCCAAGATCGCGTCCCTGCCCGACGCCGTGCCGCTCGTGCACTACAAGCTGGCGTTTCTGCGCATCGTGTGCTCCCACGAGCACTACGTGTCCCTGTCCCTTCCCGGAGGGGGCCCGGGGGGTCGCGGCGGCGCGTCCTCCCCCGCCCCCTCGGCGCACTCCGGGGGATCGAGCAGCGCGCCGCCCCCCGCCTCCGCACTCACCCACGACTTCCGGTCCCGCCACTACCTGCCCGGCCTGCTGCTGGCTGAGCTGGCCAACGCGCTGGAGATGCAGAGCCCGGTGCTGCAGAGCGCGGCGGTGGGCGCGGTGCTGGGCCTGCTCACGGCGCACGACGCGGACCCGCGCGTGGCGGAGCCCGAGCTGAAGGCGCGCGTGGCCTCGCTGTACCTGCCGCTGCTCGGCATCGTCATGGACGCCTACCCGCAGCTCTACAGGGGCTTCAACAACAACAAGGAAATCCTTCAGCTGGACAGCGAGATGAGTCAGTTCGGGAGTCTGTGCTCGCCGCCCTCCCCGGTCGACTACAAACAGGAGGGCCGCGCCGCGCTGAACAGCGAGACGAGCCGCAACCTGGTGACGTGCCTCGCCTGGGTGCTGAAGTGGGCCGGGCCCTGCGCCGCCGACCTGCCGCCCTCCGCCCTGCGCACCCTGCTCGCGCTCATCGACCTCGCCTTCAGCTGCCACGAGTACAAGGGGCGCAAGGAGATCATGAAGTGCGCTCAGCAGAACGTCCGTAAGACGACAGACATAAAGGCGAAATTAGAAGACGTCATTCTCGGCCAGGGATCGGCGCGGTCCGATTTCATCATGCGTAGGAAAGGGGGCAACATGTCCAACGGTGGGTGTAAGCGTGAGCGATGGCGCAAGGAGTGGGTGCGAGGCGGAGGGCGGGACTGCGCCCCCTCCCCCGCCGCACCGAACCCCTCGCTCGCCGCCGCGCTCGCGGGAGAGCTCTCCCTCACCCTCCTGCACGCCCTGGAGAATATCGTCCAGGCCAGCAGCAACCTGGAGTGCGGTCAGAGCATCTGCAGTACGGCGCTGCAGGTGGTGCTGCGAGCCCTGCAGCGGAACCAGAGCGTGGTAGTGCTGCAGAACATGCTGGCCACGGTGCGGGCGCTCATCGTCAGGCTGGGCTGGTCGTGCTGCGGCGAGGAGTGGAGCGTGTGCCGCGTGCTGCTGCGGCACGGCGCCGCGCTGGCGGGCGGCACGCGCGCGCACGCCGCCGCGGCGCTGTACGCGCTCATGCGCCAGCACTACCTGCTCGGCAACAACTTCAGTCGCGTGAAGATGCAGATAACGATGTCCCTGTCGTCGCTGGTGGGCACGTCCACCACCTTCAGCGAGGAGTCGCTCCGCCGCGCGCTGAAGACCATCCTGGTGTACGCCGAGCGCGACGCCGACATGCACGACACCACCTTCCCGGAGCAGGTGAAAGATCTAGTGTTCAACCTCCACATGATCCTATCGGACACGGTAAAGATGAAGGAGTACGCCGAGGACCCGGAAATGCTGCTGGATCTCATGCACCGCGTGGCGCGAGGGTACCAGCACAGCCCGGACTTGAGGCTCACCTGGCTGAACAACATGGCGCAGAAGCACATGGAGCGCTCGAACCACGCGGAGGCGGGCATGTGCCTGGTGCAGGGCGCGGCGCTGGTGGCGGAGCGGCTGGGCGCGCGCGGGCGGGGCGCGCAGCTGCTGCACCGCGTCACGCACAACGCGCTGGACGAGAGCTGCCCCGACGCGCTGCACCACCACCTCACGCACCACGAGCTGCAGGCGCTGCTGGACCACGCGGCCGGCGAGCTCATGGCGGCCGGCATGTACGAGACCGTCAACGAGCTCTACAAGGTGCTCATCCCCATCGCAGAGGAACACCGCGACTACAAGAAGCTGGCTAACATCCACAGCAAGCTGCAAGAGGCGTTCACCCGCATCGAGCAGCTGCACGGGAAGCGCGTGTTCGGGACGTACTTCCGCGTGTGCTTCTACGGGCAGCTGTTCGGGGACCTGCACCTCCAGCAGTTCGTCTACAAGGAGCACGCGCTCACCAAGCTGCCCGAGATCTTCAGCAGGCTCGAGGTCAGCCCCCGCACAACCCACCACAGTCCCATCACGAGCTTTTTTTATTGCTTAAATGGGTGGATGAGTTCACAGCCCACCTGA
Protein
MDGSCKILHEELALQWVVASGVTRDLAMQNSWALFELLVKSMSEYVQWSGAHEAARKARFPEPFTDDLTTLVNCVTSEIISGYGKNSRLTQSLNNSLAFFLFDLINIMDRGYVLSLVRTYYKQMSAKIASLPDAVPLVHYKLAFLRIVCSHEHYVSLSLPGGGPGGRGGASSPAPSAHSGGSSSAPPPASALTHDFRSRHYLPGLLLAELANALEMQSPVLQSAAVGAVLGLLTAHDADPRVAEPELKARVASLYLPLLGIVMDAYPQLYRGFNNNKEILQLDSEMSQFGSLCSPPSPVDYKQEGRAALNSETSRNLVTCLAWVLKWAGPCAADLPPSALRTLLALIDLAFSCHEYKGRKEIMKCAQQNVRKTTDIKAKLEDVILGQGSARSDFIMRRKGGNMSNGGCKRERWRKEWVRGGGRDCAPSPAAPNPSLAAALAGELSLTLLHALENIVQASSNLECGQSICSTALQVVLRALQRNQSVVVLQNMLATVRALIVRLGWSCCGEEWSVCRVLLRHGAALAGGTRAHAAAALYALMRQHYLLGNNFSRVKMQITMSLSSLVGTSTTFSEESLRRALKTILVYAERDADMHDTTFPEQVKDLVFNLHMILSDTVKMKEYAEDPEMLLDLMHRVARGYQHSPDLRLTWLNNMAQKHMERSNHAEAGMCLVQGAALVAERLGARGRGAQLLHRVTHNALDESCPDALHHHLTHHELQALLDHAAGELMAAGMYETVNELYKVLIPIAEEHRDYKKLANIHSKLQEAFTRIEQLHGKRVFGTYFRVCFYGQLFGDLHLQQFVYKEHALTKLPEIFSRLEVSPRTTHHSPITSFFYCLNGWMSSQPT
Summary
Similarity
Belongs to the DOCK family.
Uniprot
H9J946
A0A194RBG8
A0A194Q0I5
A0A1B6FQ14
A0A1B6EVF4
E0W0W5
+ More
A0A0K8VE25 A0A0K8U3G3 A0A0K8VH54 A0A0L0CG49 A0A0A1WUJ0 A0A1A9X8B6 A0A0A1XEA3 A0A1B0BZM0 A0A1B0AIS7 A0A1A9V9Z5 A0A1B0G472 W8BMR9 W8B0P4 A0A182N8K8 A0A182R6T0 A0A084W644 A0A182MU42 A0A182Q4V8 A0A182VT12 A0A182SGC5 A0A182PF79 A0A182IKN6 A0A182JPQ1 A0A1A9WAK7 A0A2M4A869 A0A2M4A9S6 A0A182XB39 A0A182V0B7 A0A1S4H392 A0A1I8QEI0 A0A182KJQ2 A0A182FQG0 A0A1I8QEK8 A0A182TK00 A0A182I0N4 Q7Q0S0 A0A1B6LB75 A0A1S4JH19 A0A2M4B8V3 A0A2M4B884 A0A1Q3G270 A0A2M3YZ18 A0A2M3YZ26 A0A1I8N762 B0WF11 A0A2M3ZFM6 A0A1I8N757 Q16R48 A0A2M4CJ19 A0A2M4CMN9 A0A147BGB9 A0A1S4FS49 A0A131YV36 A0A0T6AWK8 Q9VPI9 B4Q5L6 A0A1W4W569 A0A1W4VT38 B4ICR4 A0A0R1DQ26 B3N767 A0A0P8XUN7 B3MLR7 A0A1B6KGR3 B4P1U7 D6WNM9 A0A3B0K0R8 A0A3B0JA94 A0A0P5LNC9 A0A0R3NRA2 Q29NQ9 A0A151XIM3 A0A195CPL0 F4WYM7 A0A0C9RHM0 A0A0P5SZC3 B4GQT1 B4N781 E2AVS9 A0A026WTD7 A0A151K0V4 A0A195BTA4 A0A158NF90 A0A2J7PXZ1 A0A0P5PBF1 A0A0Q9XFN6 B4KFN0 E2BT08 A0A0P5PJX4 A0A0P5JKS1 A0A0P5SFN5 A0A0P5LPE0 A0A182GSP6 A0A3P9AKQ2
A0A0K8VE25 A0A0K8U3G3 A0A0K8VH54 A0A0L0CG49 A0A0A1WUJ0 A0A1A9X8B6 A0A0A1XEA3 A0A1B0BZM0 A0A1B0AIS7 A0A1A9V9Z5 A0A1B0G472 W8BMR9 W8B0P4 A0A182N8K8 A0A182R6T0 A0A084W644 A0A182MU42 A0A182Q4V8 A0A182VT12 A0A182SGC5 A0A182PF79 A0A182IKN6 A0A182JPQ1 A0A1A9WAK7 A0A2M4A869 A0A2M4A9S6 A0A182XB39 A0A182V0B7 A0A1S4H392 A0A1I8QEI0 A0A182KJQ2 A0A182FQG0 A0A1I8QEK8 A0A182TK00 A0A182I0N4 Q7Q0S0 A0A1B6LB75 A0A1S4JH19 A0A2M4B8V3 A0A2M4B884 A0A1Q3G270 A0A2M3YZ18 A0A2M3YZ26 A0A1I8N762 B0WF11 A0A2M3ZFM6 A0A1I8N757 Q16R48 A0A2M4CJ19 A0A2M4CMN9 A0A147BGB9 A0A1S4FS49 A0A131YV36 A0A0T6AWK8 Q9VPI9 B4Q5L6 A0A1W4W569 A0A1W4VT38 B4ICR4 A0A0R1DQ26 B3N767 A0A0P8XUN7 B3MLR7 A0A1B6KGR3 B4P1U7 D6WNM9 A0A3B0K0R8 A0A3B0JA94 A0A0P5LNC9 A0A0R3NRA2 Q29NQ9 A0A151XIM3 A0A195CPL0 F4WYM7 A0A0C9RHM0 A0A0P5SZC3 B4GQT1 B4N781 E2AVS9 A0A026WTD7 A0A151K0V4 A0A195BTA4 A0A158NF90 A0A2J7PXZ1 A0A0P5PBF1 A0A0Q9XFN6 B4KFN0 E2BT08 A0A0P5PJX4 A0A0P5JKS1 A0A0P5SFN5 A0A0P5LPE0 A0A182GSP6 A0A3P9AKQ2
Pubmed
19121390
26354079
20566863
26108605
25830018
24495485
+ More
24438588 12364791 20966253 25315136 17510324 29652888 26830274 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17550304 18362917 19820115 15632085 21719571 20798317 24508170 21347285 26483478 25069045
24438588 12364791 20966253 25315136 17510324 29652888 26830274 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17550304 18362917 19820115 15632085 21719571 20798317 24508170 21347285 26483478 25069045
EMBL
BABH01004434
BABH01004435
BABH01004436
BABH01004437
KQ460644
KPJ13246.1
+ More
KQ459584 KPI98509.1 GECZ01017473 JAS52296.1 GECZ01027793 JAS41976.1 AAZO01006860 DS235862 EEB19271.1 GDHF01015544 JAI36770.1 GDHF01031206 JAI21108.1 GDHF01030725 GDHF01014111 JAI21589.1 JAI38203.1 JRES01000438 KNC31197.1 GBXI01011573 JAD02719.1 GBXI01004538 JAD09754.1 JXJN01023184 CCAG010018118 GAMC01011889 JAB94666.1 GAMC01011890 JAB94665.1 ATLV01020721 ATLV01020722 KE525305 KFB45688.1 AXCM01000242 AXCN02001838 GGFK01003620 MBW36941.1 GGFK01004171 MBW37492.1 AAAB01008980 APCN01005224 EAA13822.4 GEBQ01019203 JAT20774.1 GGFJ01000117 MBW49258.1 GGFJ01000118 MBW49259.1 GFDL01001189 JAV33856.1 GGFM01000754 MBW21505.1 GGFM01000771 MBW21522.1 DS231913 EDS25919.1 GGFM01006534 MBW27285.1 CH477724 EAT36885.1 GGFL01001146 MBW65324.1 GGFL01001980 MBW66158.1 GEGO01005567 JAR89837.1 GEDV01006217 JAP82340.1 LJIG01022636 KRT79514.1 AE014134 BT015273 AAF51561.2 AAT94502.1 CM000361 CM002910 EDX03166.1 KMY87226.1 CH480829 EDW45340.1 CM000157 KRJ97011.1 CH954177 EDV57174.1 CH902620 KPU73033.1 EDV30788.1 GEBQ01029327 JAT10650.1 EDW87083.1 KQ971342 EFA03827.2 OUUW01000004 SPP79206.1 SPP79207.1 GDIQ01168294 JAK83431.1 CH379059 KRT03694.1 EAL34159.1 KQ982080 KYQ60138.1 KQ977444 KYN02673.1 GL888454 EGI60690.1 GBYB01012702 JAG82469.1 GDIP01133224 JAL70490.1 CH479187 EDW39953.1 CH964182 EDW80222.1 GL443213 EFN62351.1 KK107109 EZA59277.1 KQ981319 KYN42726.1 KQ976417 KYM89659.1 ADTU01014037 NEVH01020852 PNF21203.1 GDIQ01130962 JAL20764.1 CH933807 KRG02456.1 EDW10995.1 GL450325 EFN81138.1 GDIQ01129661 JAL22065.1 GDIQ01203581 JAK48144.1 GDIP01140318 JAL63396.1 GDIQ01169193 JAK82532.1 JXUM01017615 JXUM01017616 JXUM01017617 JXUM01017618 JXUM01017619 JXUM01017620 JXUM01017621 KQ560488 KXJ82146.1
KQ459584 KPI98509.1 GECZ01017473 JAS52296.1 GECZ01027793 JAS41976.1 AAZO01006860 DS235862 EEB19271.1 GDHF01015544 JAI36770.1 GDHF01031206 JAI21108.1 GDHF01030725 GDHF01014111 JAI21589.1 JAI38203.1 JRES01000438 KNC31197.1 GBXI01011573 JAD02719.1 GBXI01004538 JAD09754.1 JXJN01023184 CCAG010018118 GAMC01011889 JAB94666.1 GAMC01011890 JAB94665.1 ATLV01020721 ATLV01020722 KE525305 KFB45688.1 AXCM01000242 AXCN02001838 GGFK01003620 MBW36941.1 GGFK01004171 MBW37492.1 AAAB01008980 APCN01005224 EAA13822.4 GEBQ01019203 JAT20774.1 GGFJ01000117 MBW49258.1 GGFJ01000118 MBW49259.1 GFDL01001189 JAV33856.1 GGFM01000754 MBW21505.1 GGFM01000771 MBW21522.1 DS231913 EDS25919.1 GGFM01006534 MBW27285.1 CH477724 EAT36885.1 GGFL01001146 MBW65324.1 GGFL01001980 MBW66158.1 GEGO01005567 JAR89837.1 GEDV01006217 JAP82340.1 LJIG01022636 KRT79514.1 AE014134 BT015273 AAF51561.2 AAT94502.1 CM000361 CM002910 EDX03166.1 KMY87226.1 CH480829 EDW45340.1 CM000157 KRJ97011.1 CH954177 EDV57174.1 CH902620 KPU73033.1 EDV30788.1 GEBQ01029327 JAT10650.1 EDW87083.1 KQ971342 EFA03827.2 OUUW01000004 SPP79206.1 SPP79207.1 GDIQ01168294 JAK83431.1 CH379059 KRT03694.1 EAL34159.1 KQ982080 KYQ60138.1 KQ977444 KYN02673.1 GL888454 EGI60690.1 GBYB01012702 JAG82469.1 GDIP01133224 JAL70490.1 CH479187 EDW39953.1 CH964182 EDW80222.1 GL443213 EFN62351.1 KK107109 EZA59277.1 KQ981319 KYN42726.1 KQ976417 KYM89659.1 ADTU01014037 NEVH01020852 PNF21203.1 GDIQ01130962 JAL20764.1 CH933807 KRG02456.1 EDW10995.1 GL450325 EFN81138.1 GDIQ01129661 JAL22065.1 GDIQ01203581 JAK48144.1 GDIP01140318 JAL63396.1 GDIQ01169193 JAK82532.1 JXUM01017615 JXUM01017616 JXUM01017617 JXUM01017618 JXUM01017619 JXUM01017620 JXUM01017621 KQ560488 KXJ82146.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000009046
UP000037069
UP000092443
+ More
UP000092460 UP000092445 UP000078200 UP000092444 UP000075884 UP000075900 UP000030765 UP000075883 UP000075886 UP000075920 UP000075901 UP000075885 UP000075880 UP000075881 UP000091820 UP000076407 UP000075903 UP000095300 UP000075882 UP000069272 UP000075902 UP000075840 UP000007062 UP000095301 UP000002320 UP000008820 UP000000803 UP000000304 UP000192221 UP000001292 UP000002282 UP000008711 UP000007801 UP000007266 UP000268350 UP000001819 UP000075809 UP000078542 UP000007755 UP000008744 UP000007798 UP000000311 UP000053097 UP000078541 UP000078540 UP000005205 UP000235965 UP000009192 UP000008237 UP000069940 UP000249989 UP000265140
UP000092460 UP000092445 UP000078200 UP000092444 UP000075884 UP000075900 UP000030765 UP000075883 UP000075886 UP000075920 UP000075901 UP000075885 UP000075880 UP000075881 UP000091820 UP000076407 UP000075903 UP000095300 UP000075882 UP000069272 UP000075902 UP000075840 UP000007062 UP000095301 UP000002320 UP000008820 UP000000803 UP000000304 UP000192221 UP000001292 UP000002282 UP000008711 UP000007801 UP000007266 UP000268350 UP000001819 UP000075809 UP000078542 UP000007755 UP000008744 UP000007798 UP000000311 UP000053097 UP000078541 UP000078540 UP000005205 UP000235965 UP000009192 UP000008237 UP000069940 UP000249989 UP000265140
Pfam
Interpro
IPR027357
DHR-2
+ More
IPR037808 C2_Dock-C
IPR021816 DOCK_C/D_N
IPR010703 DOCK_C
IPR026791 DOCK
IPR035892 C2_domain_sf
IPR027007 DHR-1_domain
IPR016024 ARM-type_fold
IPR032376 DOCK_N
IPR002049 Laminin_EGF
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR018097 EGF_Ca-bd_CS
IPR018247 EF_Hand_1_Ca_BS
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR037808 C2_Dock-C
IPR021816 DOCK_C/D_N
IPR010703 DOCK_C
IPR026791 DOCK
IPR035892 C2_domain_sf
IPR027007 DHR-1_domain
IPR016024 ARM-type_fold
IPR032376 DOCK_N
IPR002049 Laminin_EGF
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR018097 EGF_Ca-bd_CS
IPR018247 EF_Hand_1_Ca_BS
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
Gene 3D
ProteinModelPortal
H9J946
A0A194RBG8
A0A194Q0I5
A0A1B6FQ14
A0A1B6EVF4
E0W0W5
+ More
A0A0K8VE25 A0A0K8U3G3 A0A0K8VH54 A0A0L0CG49 A0A0A1WUJ0 A0A1A9X8B6 A0A0A1XEA3 A0A1B0BZM0 A0A1B0AIS7 A0A1A9V9Z5 A0A1B0G472 W8BMR9 W8B0P4 A0A182N8K8 A0A182R6T0 A0A084W644 A0A182MU42 A0A182Q4V8 A0A182VT12 A0A182SGC5 A0A182PF79 A0A182IKN6 A0A182JPQ1 A0A1A9WAK7 A0A2M4A869 A0A2M4A9S6 A0A182XB39 A0A182V0B7 A0A1S4H392 A0A1I8QEI0 A0A182KJQ2 A0A182FQG0 A0A1I8QEK8 A0A182TK00 A0A182I0N4 Q7Q0S0 A0A1B6LB75 A0A1S4JH19 A0A2M4B8V3 A0A2M4B884 A0A1Q3G270 A0A2M3YZ18 A0A2M3YZ26 A0A1I8N762 B0WF11 A0A2M3ZFM6 A0A1I8N757 Q16R48 A0A2M4CJ19 A0A2M4CMN9 A0A147BGB9 A0A1S4FS49 A0A131YV36 A0A0T6AWK8 Q9VPI9 B4Q5L6 A0A1W4W569 A0A1W4VT38 B4ICR4 A0A0R1DQ26 B3N767 A0A0P8XUN7 B3MLR7 A0A1B6KGR3 B4P1U7 D6WNM9 A0A3B0K0R8 A0A3B0JA94 A0A0P5LNC9 A0A0R3NRA2 Q29NQ9 A0A151XIM3 A0A195CPL0 F4WYM7 A0A0C9RHM0 A0A0P5SZC3 B4GQT1 B4N781 E2AVS9 A0A026WTD7 A0A151K0V4 A0A195BTA4 A0A158NF90 A0A2J7PXZ1 A0A0P5PBF1 A0A0Q9XFN6 B4KFN0 E2BT08 A0A0P5PJX4 A0A0P5JKS1 A0A0P5SFN5 A0A0P5LPE0 A0A182GSP6 A0A3P9AKQ2
A0A0K8VE25 A0A0K8U3G3 A0A0K8VH54 A0A0L0CG49 A0A0A1WUJ0 A0A1A9X8B6 A0A0A1XEA3 A0A1B0BZM0 A0A1B0AIS7 A0A1A9V9Z5 A0A1B0G472 W8BMR9 W8B0P4 A0A182N8K8 A0A182R6T0 A0A084W644 A0A182MU42 A0A182Q4V8 A0A182VT12 A0A182SGC5 A0A182PF79 A0A182IKN6 A0A182JPQ1 A0A1A9WAK7 A0A2M4A869 A0A2M4A9S6 A0A182XB39 A0A182V0B7 A0A1S4H392 A0A1I8QEI0 A0A182KJQ2 A0A182FQG0 A0A1I8QEK8 A0A182TK00 A0A182I0N4 Q7Q0S0 A0A1B6LB75 A0A1S4JH19 A0A2M4B8V3 A0A2M4B884 A0A1Q3G270 A0A2M3YZ18 A0A2M3YZ26 A0A1I8N762 B0WF11 A0A2M3ZFM6 A0A1I8N757 Q16R48 A0A2M4CJ19 A0A2M4CMN9 A0A147BGB9 A0A1S4FS49 A0A131YV36 A0A0T6AWK8 Q9VPI9 B4Q5L6 A0A1W4W569 A0A1W4VT38 B4ICR4 A0A0R1DQ26 B3N767 A0A0P8XUN7 B3MLR7 A0A1B6KGR3 B4P1U7 D6WNM9 A0A3B0K0R8 A0A3B0JA94 A0A0P5LNC9 A0A0R3NRA2 Q29NQ9 A0A151XIM3 A0A195CPL0 F4WYM7 A0A0C9RHM0 A0A0P5SZC3 B4GQT1 B4N781 E2AVS9 A0A026WTD7 A0A151K0V4 A0A195BTA4 A0A158NF90 A0A2J7PXZ1 A0A0P5PBF1 A0A0Q9XFN6 B4KFN0 E2BT08 A0A0P5PJX4 A0A0P5JKS1 A0A0P5SFN5 A0A0P5LPE0 A0A182GSP6 A0A3P9AKQ2
PDB
2WMO
E-value=1.0076e-27,
Score=310
Ontologies
PANTHER
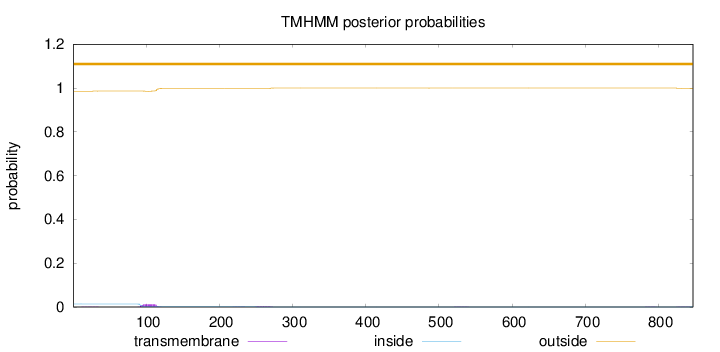
Topology
Length:
847
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.34803
Exp number, first 60 AAs:
0.0141
Total prob of N-in:
0.01414
outside
1 - 847
Population Genetic Test Statistics
Pi
259.454226
Theta
164.171394
Tajima's D
1.374642
CLR
0.45376
CSRT
0.75761211939403
Interpretation
Uncertain
