Gene
KWMTBOMO01674
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.629 Nuclear Reliability : 1.385
Sequence
CDS
ATGACATTTGGCGCGTGGAACGTGAGAACGCTTCTTGATCGAGATGGCAACGCCTGCCCTGAACGCAAGACCGCCATAGTAGCTCGGGAACTTCATCGCTACAACGTAGATGTGGCTGCTCTCAGCGAAACACATCTTGCCGATGAAGGAGAGCTGGTGGAAGTAGGTGCTGGTTATACTTTCTTCTGGAAAGGTACCGCTGCCTCTGAAACGCGACACTCAGGTGTAGGATTTGCCATCAAGAATCACTTAGTAAAGCGATTAGAGGAGTACCCTGTACATATCTCGGACCGCGTTACCACACTGCGAGTTCACCTGGATAAAGACAATTACCTTAATGTCATCAGTGTATATGCTCCAACGCTTGACAAGTCTGATGACATTAAGGACAAATTCTATGGGGAAGTGACTCTTTGCCTTGACAGTATAAACGCCAGAGAGCAGGTACTATTGCTGGGCGACTTCAATGCCAGGGTTGGTCGGGACTATGAGGCTTGGCCTGGAGTTCTGGGTAGACACGGAGTCGGCAACATGAACAGCAATGGTCAGTTGCTGCTCAGTCTTTGTGCTCAATACGGTCTAGCAATTACGAACACTATGTTTAGACTTGCCGCTAAGTACAAGACAACATGGATGCACCCAAGATCCAAGCACTGGCATTTGATCGACTATGCTATCGTAAGGCAAAGAGATTTCAGCCAAGTGCAGATCACCCGTGTTATGCGTGGTGCGCACTGCTGGTCTGACCACCGACTTATTGTCACTAAGCTACGACTCCGCCTCCGCCGCCCGCGTAGATCTCCTATGAAAAAGCTTGCGTCTTTGGACATAGAGAAGCTAAGAGATCCTGAGGTGAGGAAGAACTATGCTGAAGTTTTGTCTAAGAAGCTGTTGCCAGTTAATGAGACAGATGATGTTGATGCTGACTGGGGAGCTTTATCATCTCATATCATGGATACCGCTTCAATTGCACTGGGTAGGAAATGCCGTCGTAATGAAGACTGGTTTGATGGAAATGATGAAGTTTTGCGGGAGGCACTCGACAAACACCGTGATCTCTTGCGACAGCACAGAAGGCGTAAAGGAAGCGTTGCGGCGGTCAAAGCTAGTGACCTTGAACTGCGCAAGCTGTCTCGACAAATAAAAGACAAGTGGTGGCAGGACAAAGCTATTCATATGCAATGGCTCACTGACACAAACCAGCTCGGTGAGTTCTATAGCGAGGTGCGCAAGCTGATGGGTACATCCAACCATGCAAAGGTTCCTTTAAGGGCTCTAGATAGTAGGCACCTCTTAACAAGCAAGGAGGATGTACTAAGGCGCTGGGCAGAGCACTTCAATGCCCTGCTGAATGTGGATCGATCAGCAGATCTGCAAAGCATCGCTTTAATGCCTCAACTCCCTCTTGCCCTTGAGCTGGACGAGCCCCTATTGCGTGACGAGGTTGTTACTGCCATCAGACAGCAAAAAAATAAAAGGGCGGCTGGCGCTGACCTTATACCAGGAGAGCTGATCAAGTACGGTGGAGAGGATTTGCACACGTTGGTTTGGGAACTGTTTGTTCGTATGTGGGAGGAAGAGCGCGTTCCGGATAGCTTTAAAGTATCGCGCATAACTGCTCTCTATAAGAACAGGGGCGACCGATCTGACTGCGACTCCTACCGCGGTATTTCGCTCCTATCGGCCCCCGGAAAGGTCTTTGCTAGGGTACTCTTAAACCGCCTAAAGGACTTATCTGAAAAGATCCTGCCCGAAACTCAGTTTGGCTTTCGACCGGATCGAGGCACATGCGAAGCGATCTTCTCTGTGCGTCAACTACAAGAAAAGAGCAGAGAACAGGGTCGTCAGCTGTACCTCTGTTTTGTTGACCTGGAGAAGGCTTTTGACAGCGTGCCTCGCGAAGCCTTATGGTTGGTGTTACGGAAGCTCGGATGCACAGAAAAGTTTGTGGCGCTTCTTAGACTCCTGCATGACGACATGCAATGCTGCGTCACTGTAGATGGTGAACAGACGGGCTTCTTCCCCGTTACCTGTGGGGTGAAGCAAGGATGCGTCTTAGCGCCTACTCTGTTCGCCTTGTACTTTGCGGTCGTAGTTAGAGAGGTTCTACAAACTGTTTCCCAAGGAGTCCGCATCCGCTTCCGTACGGACGGCAGCCTTTTCAATTTGGCTAGACTTAAGGCTCGCACCCAAGTGTCGTATGCATTGATCACTGAGATCATTTATGCTGATGACTTGTGCTTCTTAGCAGAATCCCCAGACGGCCTGCAACAGCTGATGTCCGTTTTTCACCAAGCCTGCCGTAAATTCGGCCTGAAGATTAGTGTCGATAAGACGGAGGTGATGTCCCTGGACAGTCACGGTCACGAGACACTAACCATAAAGCTTGGTGAGGATGTGTTGAAGCAGGTGGATAAATATCGCTATCTGGGGAACACTTTAACATCCAAGTGTGACCTTGATGATGAGATCAATAGCAGGGTCGGTGCCGCTGCGGCAGCATTTGGAAAGCTATATTCCAAGGTCTTCTGCTCACACGACCTTAAGTTGGCCACTAAAATCTCAGTTTACATGGCAATTGTGCTGCCAAGCCTCCTATACTCCTCCGAGTCGTGGTGTGTGTACCGCCGCCACATTCGCACTCTGGATCGCTTCCACCTTAAATGCCTTCGTGCCATCATGAAAATCAGATGGTCTGATAGAGTGCGAAATACCGAAGTCTTGCGACGTGCCAATGTTGGTGGCATTGAGACCTATTTAATGCGCCGGCAGCTTCGATGGTGCGGTCATGTATCACGCATGACGGAGGAAAGAGTGGCGAAGCGCATCTTCTTCTCTGAATTGCAGGATGGCAAGCGAAAGCATGGCGGACAACTCCTGCGGTACAAGGATGTCGTGAAACGACACATGAAGAGATGTGATATAGAGCCCTCTCAATGGGAGCGCTTGGCAGCACAGCGGCCAGAATGGCGCAGGATGGTGAACAGCAAAGTACGCGAGTTCGAGGATCAGCGTAAAGCTGACCTTGATTACAAACGCGACCAGTTGAAGGCCCGCCCACCTGCTGCCATAACCTATAATTATGAGAACGGCGTGCTTACGTGCCCTCAATGCGCAAGGAGTTTTGCCGCAAAGATAGGCTATATAAGCCATCTTCGAGCGCACGAGCGCCAAATCGACGGATAG
Protein
MTFGAWNVRTLLDRDGNACPERKTAIVARELHRYNVDVAALSETHLADEGELVEVGAGYTFFWKGTAASETRHSGVGFAIKNHLVKRLEEYPVHISDRVTTLRVHLDKDNYLNVISVYAPTLDKSDDIKDKFYGEVTLCLDSINAREQVLLLGDFNARVGRDYEAWPGVLGRHGVGNMNSNGQLLLSLCAQYGLAITNTMFRLAAKYKTTWMHPRSKHWHLIDYAIVRQRDFSQVQITRVMRGAHCWSDHRLIVTKLRLRLRRPRRSPMKKLASLDIEKLRDPEVRKNYAEVLSKKLLPVNETDDVDADWGALSSHIMDTASIALGRKCRRNEDWFDGNDEVLREALDKHRDLLRQHRRRKGSVAAVKASDLELRKLSRQIKDKWWQDKAIHMQWLTDTNQLGEFYSEVRKLMGTSNHAKVPLRALDSRHLLTSKEDVLRRWAEHFNALLNVDRSADLQSIALMPQLPLALELDEPLLRDEVVTAIRQQKNKRAAGADLIPGELIKYGGEDLHTLVWELFVRMWEEERVPDSFKVSRITALYKNRGDRSDCDSYRGISLLSAPGKVFARVLLNRLKDLSEKILPETQFGFRPDRGTCEAIFSVRQLQEKSREQGRQLYLCFVDLEKAFDSVPREALWLVLRKLGCTEKFVALLRLLHDDMQCCVTVDGEQTGFFPVTCGVKQGCVLAPTLFALYFAVVVREVLQTVSQGVRIRFRTDGSLFNLARLKARTQVSYALITEIIYADDLCFLAESPDGLQQLMSVFHQACRKFGLKISVDKTEVMSLDSHGHETLTIKLGEDVLKQVDKYRYLGNTLTSKCDLDDEINSRVGAAAAAFGKLYSKVFCSHDLKLATKISVYMAIVLPSLLYSSESWCVYRRHIRTLDRFHLKCLRAIMKIRWSDRVRNTEVLRRANVGGIETYLMRRQLRWCGHVSRMTEERVAKRIFFSELQDGKRKHGGQLLRYKDVVKRHMKRCDIEPSQWERLAAQRPEWRRMVNSKVREFEDQRKADLDYKRDQLKARPPAAITYNYENGVLTCPQCARSFAAKIGYISHLRAHERQIDG
Summary
Uniprot
D7F171
A0A3P9H393
A0A1B3B7F1
K7EXX0
K7EZH1
K7F1H9
+ More
A0A3B3I759 A0A3B3HTA8 A0A0B7BSR0 K7EZE2 A0A3P9JDD8 W4YWJ6 K7EWE4 A0A0X3PCE5 A0A2B4RCM2 H2ZZE9 A0A3B3HUN9 A0A2B4SPJ1 H3B828 A0A146SQ82 A0A146P9F6 A0A3B5Q8W9 A0A2B4R8E3 A0A2B4RLZ4 A0A3B5R2H7 A0A3B5QX16 A0A3B5QFT5 A0A3P9PV83 A0A0G4GUQ4 A0A2B7ZXS2 A0A1Y1JWU5 C7C207 D7F173 Q4QQE8 A0A2S2PT92 A0A1Y1KJ02 A0A3P9LYS6 A0A1Y1LZ08 A0A060Q6A2 A0A2P8ZFZ0 A0A2P8ZLT6 A0A2P8Z488 A0A2P8Z313 A0A3P9M1B5 A0A2P8YWA2 A0A023EX67 A0A2P8Y2S1 A0A1Y1MCV0 A0A016TPK0 A0A224XIY2 A0A2P8YTB7 A0A2P8XC08 J9KNQ8 A0A2M4ACI4 A0A2M4A809 A0A1B0DRK3 A0A3B3H8A8 A0A2M4A2X8 A0A023F0F3
A0A3B3I759 A0A3B3HTA8 A0A0B7BSR0 K7EZE2 A0A3P9JDD8 W4YWJ6 K7EWE4 A0A0X3PCE5 A0A2B4RCM2 H2ZZE9 A0A3B3HUN9 A0A2B4SPJ1 H3B828 A0A146SQ82 A0A146P9F6 A0A3B5Q8W9 A0A2B4R8E3 A0A2B4RLZ4 A0A3B5R2H7 A0A3B5QX16 A0A3B5QFT5 A0A3P9PV83 A0A0G4GUQ4 A0A2B7ZXS2 A0A1Y1JWU5 C7C207 D7F173 Q4QQE8 A0A2S2PT92 A0A1Y1KJ02 A0A3P9LYS6 A0A1Y1LZ08 A0A060Q6A2 A0A2P8ZFZ0 A0A2P8ZLT6 A0A2P8Z488 A0A2P8Z313 A0A3P9M1B5 A0A2P8YWA2 A0A023EX67 A0A2P8Y2S1 A0A1Y1MCV0 A0A016TPK0 A0A224XIY2 A0A2P8YTB7 A0A2P8XC08 J9KNQ8 A0A2M4ACI4 A0A2M4A809 A0A1B0DRK3 A0A3B3H8A8 A0A2M4A2X8 A0A023F0F3
Pubmed
EMBL
FJ265556
ADI61824.1
KU315381
AOE48155.1
AGCU01074235
AGCU01019305
+ More
AGCU01078395 AGCU01195602 HACG01049402 HACG01049403 HACG01049405 HACG01049409 CEK96267.1 CEK96268.1 CEK96270.1 CEK96274.1 AGCU01099253 AAGJ04079542 AGCU01159961 GEEE01015618 JAP47607.1 LSMT01000835 PFX14128.1 AFYH01233515 LSMT01000050 PFX30432.1 AFYH01086022 GCES01103567 JAQ82755.1 GCES01145874 JAQ40448.1 LSMT01001244 PFX12648.1 LSMT01000478 PFX17357.1 CDMZ01001569 CEM34549.1 PDHN01000949 PGH37547.1 GEZM01098732 JAV53782.1 FN356221 CAX83710.1 FJ265558 ADI61826.1 BN000794 CAJ00236.1 GGMR01020071 MBY32690.1 GEZM01082534 GEZM01082533 JAV61334.1 GEZM01043748 JAV78864.1 HE799691 CCH14900.1 PYGN01000070 PSN55417.1 PYGN01000020 PSN57453.1 PYGN01000203 PSN51319.1 PYGN01000219 PSN50892.1 PYGN01000324 PSN48495.1 GAPW01000107 JAC13491.1 PYGN01001004 PSN38555.1 GEZM01039814 JAV81167.1 JARK01001422 EYC04705.1 GFTR01008437 JAW07989.1 PYGN01000372 PSN47498.1 PYGN01003402 PSN29545.1 ABLF02039015 GGFK01005184 MBW38505.1 GGFK01003613 MBW36934.1 AJVK01001578 GGFK01001798 MBW35119.1 GBBI01004261 JAC14451.1
AGCU01078395 AGCU01195602 HACG01049402 HACG01049403 HACG01049405 HACG01049409 CEK96267.1 CEK96268.1 CEK96270.1 CEK96274.1 AGCU01099253 AAGJ04079542 AGCU01159961 GEEE01015618 JAP47607.1 LSMT01000835 PFX14128.1 AFYH01233515 LSMT01000050 PFX30432.1 AFYH01086022 GCES01103567 JAQ82755.1 GCES01145874 JAQ40448.1 LSMT01001244 PFX12648.1 LSMT01000478 PFX17357.1 CDMZ01001569 CEM34549.1 PDHN01000949 PGH37547.1 GEZM01098732 JAV53782.1 FN356221 CAX83710.1 FJ265558 ADI61826.1 BN000794 CAJ00236.1 GGMR01020071 MBY32690.1 GEZM01082534 GEZM01082533 JAV61334.1 GEZM01043748 JAV78864.1 HE799691 CCH14900.1 PYGN01000070 PSN55417.1 PYGN01000020 PSN57453.1 PYGN01000203 PSN51319.1 PYGN01000219 PSN50892.1 PYGN01000324 PSN48495.1 GAPW01000107 JAC13491.1 PYGN01001004 PSN38555.1 GEZM01039814 JAV81167.1 JARK01001422 EYC04705.1 GFTR01008437 JAW07989.1 PYGN01000372 PSN47498.1 PYGN01003402 PSN29545.1 ABLF02039015 GGFK01005184 MBW38505.1 GGFK01003613 MBW36934.1 AJVK01001578 GGFK01001798 MBW35119.1 GBBI01004261 JAC14451.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F171
A0A3P9H393
A0A1B3B7F1
K7EXX0
K7EZH1
K7F1H9
+ More
A0A3B3I759 A0A3B3HTA8 A0A0B7BSR0 K7EZE2 A0A3P9JDD8 W4YWJ6 K7EWE4 A0A0X3PCE5 A0A2B4RCM2 H2ZZE9 A0A3B3HUN9 A0A2B4SPJ1 H3B828 A0A146SQ82 A0A146P9F6 A0A3B5Q8W9 A0A2B4R8E3 A0A2B4RLZ4 A0A3B5R2H7 A0A3B5QX16 A0A3B5QFT5 A0A3P9PV83 A0A0G4GUQ4 A0A2B7ZXS2 A0A1Y1JWU5 C7C207 D7F173 Q4QQE8 A0A2S2PT92 A0A1Y1KJ02 A0A3P9LYS6 A0A1Y1LZ08 A0A060Q6A2 A0A2P8ZFZ0 A0A2P8ZLT6 A0A2P8Z488 A0A2P8Z313 A0A3P9M1B5 A0A2P8YWA2 A0A023EX67 A0A2P8Y2S1 A0A1Y1MCV0 A0A016TPK0 A0A224XIY2 A0A2P8YTB7 A0A2P8XC08 J9KNQ8 A0A2M4ACI4 A0A2M4A809 A0A1B0DRK3 A0A3B3H8A8 A0A2M4A2X8 A0A023F0F3
A0A3B3I759 A0A3B3HTA8 A0A0B7BSR0 K7EZE2 A0A3P9JDD8 W4YWJ6 K7EWE4 A0A0X3PCE5 A0A2B4RCM2 H2ZZE9 A0A3B3HUN9 A0A2B4SPJ1 H3B828 A0A146SQ82 A0A146P9F6 A0A3B5Q8W9 A0A2B4R8E3 A0A2B4RLZ4 A0A3B5R2H7 A0A3B5QX16 A0A3B5QFT5 A0A3P9PV83 A0A0G4GUQ4 A0A2B7ZXS2 A0A1Y1JWU5 C7C207 D7F173 Q4QQE8 A0A2S2PT92 A0A1Y1KJ02 A0A3P9LYS6 A0A1Y1LZ08 A0A060Q6A2 A0A2P8ZFZ0 A0A2P8ZLT6 A0A2P8Z488 A0A2P8Z313 A0A3P9M1B5 A0A2P8YWA2 A0A023EX67 A0A2P8Y2S1 A0A1Y1MCV0 A0A016TPK0 A0A224XIY2 A0A2P8YTB7 A0A2P8XC08 J9KNQ8 A0A2M4ACI4 A0A2M4A809 A0A1B0DRK3 A0A3B3H8A8 A0A2M4A2X8 A0A023F0F3
PDB
6AR3
E-value=0.0822626,
Score=88
Ontologies
GO
PANTHER
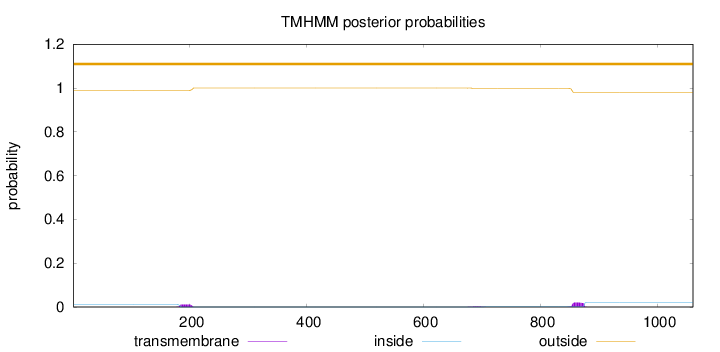
Topology
Length:
1061
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.684060000000001
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.01136
outside
1 - 1061
Population Genetic Test Statistics
Pi
36.794631
Theta
3.133118
Tajima's D
0.430839
CLR
26.407888
CSRT
0.534573271336433
Interpretation
Uncertain
