Pre Gene Modal
BGIBMGA006108
Annotation
prolactin_regulatory_binding-element_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.893
Sequence
CDS
ATGTCTCCTCATAGGTCTGATGGATTATTAGCTAAAGTTGATTTTCCACTTTACACGCTCCAAACCTTAACAAATCGACATGTCATTGTTGCTGGAGGAGGAGGAGCATCGAACACAGGTGTCGCAAATGGATTTGAAATATTTGAACTGTCACATAATGGGAAGAGGTTTGTTGCTGAAGAGGTGATGCGCCATGAGACTGGTCCTAATGTCGTGATGACGTGTTCCGTAAGGAGCATCCAGAACCGGACGTACCTCACAGCGGGACAAGAGAGCCACTGTCAGCTGTACAAGGTGAACATCCGCATGGTGGATGCTGCTGAAATGAGACGAGGAAGTTTCCGGGCTGAGAATGGGCTTGTGAGGCGGAGGAGACGGACAGTCTCTGAAAATGATAATATATCAAAGAAAAACAATGTTAGTAACACAGAGAAAAGAATGTCATTTGAAATCAGACCTTGTGATAGTGTGCAGACTGACTTTAGTAATGATCCACTGCAGCGTGTTGTCAGGATTAGTAATAATGGGAAGTTGATGGCTACAGGTGGCATCGACACCAAGGTCAGAGTTTGGACCTTTCCAAAAATGGAACTCCTTTACGTACTGGAAAAGCATACTAAAGAGTTGGACGATCTAGACTTCAGTCCGTGCGATACGACGTTAGTGAGCATCGGGAAGGACGGTCTCGCCTGCCTGTGGTCCACTGCGGGCCTGCGCCCCCCGCTCCTCACCCTCACGTGCCAGCCACCTAACGGGAACAAGTACCTCTTCAGACGTTGCAGGTTCGGGCCCAACGAAGACAAGGAAGACGCGCACAGAATGTTCACGATAGCGAACCCGCTGAGTCGCTCCGGCAAACAGAAAGGTCTGCTGCAGCTCTGGGACCCCAAGGACGGCTCGCTGCGGAAGACGGTGCTGGTGAACGAGTCTCTGTCGGCGCTGACCGTCCGGGACGACGGCCGCTTCGTGGGCGTCGGGACCATGTTCAGTGGCTCCGTCGACATTTACATCGCCTTCAGTTTGCAGCGCGTGCTGCACGTGAGGAGCGCGCACCGGATGTTCGTGACGGGCGTGCAGTTCCTGCCGGTGCGCGGGTACGGGCCCGCCGTGGCCAGCCGCAGCGAGGCCGCGCTGCTCAGCATCTCCGTCGACAACTGCCTCTGCGTGCACAGCCTGCCCTACCGCGGCTCCGTGCCCATCTGGATCGCCATCGTTCTCATCATCTTCGTACTATTCTGTACGTTCAGTCTATGTTCATATTTAGGTATTTAA
Protein
MSPHRSDGLLAKVDFPLYTLQTLTNRHVIVAGGGGASNTGVANGFEIFELSHNGKRFVAEEVMRHETGPNVVMTCSVRSIQNRTYLTAGQESHCQLYKVNIRMVDAAEMRRGSFRAENGLVRRRRRTVSENDNISKKNNVSNTEKRMSFEIRPCDSVQTDFSNDPLQRVVRISNNGKLMATGGIDTKVRVWTFPKMELLYVLEKHTKELDDLDFSPCDTTLVSIGKDGLACLWSTAGLRPPLLTLTCQPPNGNKYLFRRCRFGPNEDKEDAHRMFTIANPLSRSGKQKGLLQLWDPKDGSLRKTVLVNESLSALTVRDDGRFVGVGTMFSGSVDIYIAFSLQRVLHVRSAHRMFVTGVQFLPVRGYGPAVASRSEAALLSISVDNCLCVHSLPYRGSVPIWIAIVLIIFVLFCTFSLCSYLGI
Summary
Uniprot
H9J9B6
Q1HQ78
A0A2W1BGI8
A0A2A4IZ04
A0A2H1VXU2
S4PKN2
+ More
A0A212EWJ1 A0A0L7KZ05 A0A1L8DVU4 A0A0K8TTU7 A0A1B0D0S7 B0WE33 A0A1Q3F4K2 A0A182JN74 A0A084WG22 A0A182K7U2 A0A182MR26 A0A182HRE2 A0A182NAV9 A0A2M4BN61 Q7Q168 A0A182V0D8 A0A182KYZ7 A0A182UFT3 A0A2J7QAZ4 A0A182PG50 A0A0K8VSB4 A0A182SHZ7 A0A182XHT4 A0A182Y6E5 A0A182F4Q4 W5J6Y9 W8C8M4 A0A182RRB9 A0A182W8X1 Q16KX6 A0A034W887 A0A182QV26 A0A1Y1KNU1 A0A0A1WNB4 A0A336MKY6 U5EWI6 A0A1I8MZN2 B3N5F9 B4I1R8 B4Q4J0 A0A336N041 A0A1I8QAQ8 A0A1W4VAT6 B4KL84 B4P0P0 Q9VMH4 Q95SQ7 B3MKH8 Q29PP5 A0A3B0KM02 B4LRM8 A0A2M4AN82 B4G6H9 B4JB00 A0A0L0BW54 A0A0M5J8U9 A0A0M4ENX0 A0A1J1IH73 A0A1B6E7Q3 A0A182HC93 A0A069DYZ2 B4MUW1 A0A0V0G349 A0A224XNX5 A0A067RJH6 A0A1A9V1A3 A0A1W4WWP5 A0A1B0A875 A0A1B0BLT1 A0A1A9Y7J3 A0A1B0G0U4 A0A2J7QB04 N6TDI8 R4FNS0 A0A0P4VU66 A0A310SU50 A0A0L7QVM4 A0A0A9Y3A6 D6WP28 A0A2A3ENR3 A0A026WU94 A0A1B6LG88 A0A1B6DQD1 A0A088A1C2 F4WJM3 A0A170Z2L7 A0A0J7NUD4 A0A2P8ZJR5 A0A158NNC0 A0A195FY01 A0A0N0U4Q0 A0A195E4K8
A0A212EWJ1 A0A0L7KZ05 A0A1L8DVU4 A0A0K8TTU7 A0A1B0D0S7 B0WE33 A0A1Q3F4K2 A0A182JN74 A0A084WG22 A0A182K7U2 A0A182MR26 A0A182HRE2 A0A182NAV9 A0A2M4BN61 Q7Q168 A0A182V0D8 A0A182KYZ7 A0A182UFT3 A0A2J7QAZ4 A0A182PG50 A0A0K8VSB4 A0A182SHZ7 A0A182XHT4 A0A182Y6E5 A0A182F4Q4 W5J6Y9 W8C8M4 A0A182RRB9 A0A182W8X1 Q16KX6 A0A034W887 A0A182QV26 A0A1Y1KNU1 A0A0A1WNB4 A0A336MKY6 U5EWI6 A0A1I8MZN2 B3N5F9 B4I1R8 B4Q4J0 A0A336N041 A0A1I8QAQ8 A0A1W4VAT6 B4KL84 B4P0P0 Q9VMH4 Q95SQ7 B3MKH8 Q29PP5 A0A3B0KM02 B4LRM8 A0A2M4AN82 B4G6H9 B4JB00 A0A0L0BW54 A0A0M5J8U9 A0A0M4ENX0 A0A1J1IH73 A0A1B6E7Q3 A0A182HC93 A0A069DYZ2 B4MUW1 A0A0V0G349 A0A224XNX5 A0A067RJH6 A0A1A9V1A3 A0A1W4WWP5 A0A1B0A875 A0A1B0BLT1 A0A1A9Y7J3 A0A1B0G0U4 A0A2J7QB04 N6TDI8 R4FNS0 A0A0P4VU66 A0A310SU50 A0A0L7QVM4 A0A0A9Y3A6 D6WP28 A0A2A3ENR3 A0A026WU94 A0A1B6LG88 A0A1B6DQD1 A0A088A1C2 F4WJM3 A0A170Z2L7 A0A0J7NUD4 A0A2P8ZJR5 A0A158NNC0 A0A195FY01 A0A0N0U4Q0 A0A195E4K8
Pubmed
19121390
28756777
23622113
22118469
26227816
26369729
+ More
24438588 12364791 14747013 17210077 20966253 25244985 20920257 23761445 24495485 17510324 25348373 28004739 25830018 25315136 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 26108605 26483478 26334808 24845553 23537049 27129103 25401762 26823975 18362917 19820115 24508170 30249741 21719571 29403074 21347285
24438588 12364791 14747013 17210077 20966253 25244985 20920257 23761445 24495485 17510324 25348373 28004739 25830018 25315136 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 26108605 26483478 26334808 24845553 23537049 27129103 25401762 26823975 18362917 19820115 24508170 30249741 21719571 29403074 21347285
EMBL
BABH01004424
DQ443174
ABF51263.1
KZ150155
PZC72794.1
NWSH01004520
+ More
PCG64991.1 ODYU01005101 SOQ45643.1 GAIX01004400 JAA88160.1 AGBW02011959 OWR45868.1 JTDY01004323 KOB68296.1 GFDF01003523 JAV10561.1 GDAI01000035 JAI17568.1 AJVK01021645 DS231904 EDS45209.1 GFDL01012596 JAV22449.1 ATLV01023433 KE525343 KFB49166.1 AXCM01005445 APCN01001695 GGFJ01005376 MBW54517.1 AAAB01008980 EAA14036.3 NEVH01016304 PNF25753.1 GDHF01016458 GDHF01010834 GDHF01009540 GDHF01004598 JAI35856.1 JAI41480.1 JAI42774.1 JAI47716.1 ADMH02001962 ETN60222.1 GAMC01002909 JAC03647.1 CH477933 EAT34957.1 GAKP01008961 GAKP01008960 GAKP01008959 JAC49993.1 AXCN02000654 GEZM01080460 JAV62131.1 GBXI01014394 JAC99897.1 UFQT01001122 SSX29027.1 GANO01000552 JAB59319.1 CH954177 EDV59038.1 CH480820 EDW54475.1 CM000361 CM002910 EDX03917.1 KMY88466.1 UFQT01003842 SSX35305.1 CH933807 EDW11745.1 CM000157 EDW87935.1 AE014134 BT044528 AAF52344.1 ACH95302.1 AY060647 AAL28195.1 CH902620 EDV31531.1 CH379058 EAL34247.1 OUUW01000010 SPP86121.1 CH940649 EDW63623.1 GGFK01008910 MBW42231.1 CH479180 EDW28204.1 CH916368 EDW02870.1 JRES01001249 KNC24246.1 CP012523 ALC38368.1 ALC38182.1 CVRI01000047 CRK97785.1 GEDC01003327 JAS33971.1 JXUM01126916 JXUM01126917 JXUM01126918 KQ567125 KXJ69610.1 GBGD01001545 JAC87344.1 CH963857 EDW76306.1 GECL01003599 JAP02525.1 GFTR01006154 JAW10272.1 KK852432 KDR23972.1 JXJN01016549 CCAG010006198 PNF25754.1 APGK01006704 APGK01034104 KB740904 KB736898 KB632006 ENN78434.1 ENN83113.1 ERL87945.1 ACPB03022604 GAHY01000468 JAA77042.1 GDKW01000388 JAI56207.1 KQ760343 OAD60859.1 KQ414725 KOC62665.1 GBHO01016970 GBRD01001301 GDHC01016797 GDHC01001501 JAG26634.1 JAG64520.1 JAQ01832.1 JAQ17128.1 KQ971343 EFA04425.1 KZ288215 PBC32769.1 KK107109 QOIP01000008 EZA59226.1 RLU19695.1 GEBQ01017339 JAT22638.1 GEDC01009403 JAS27895.1 GL888186 EGI65515.1 GEMB01002673 JAS00520.1 LBMM01001611 KMQ96015.1 PYGN01000035 PSN56741.1 ADTU01021228 KQ981208 KYN44714.1 KQ435803 KOX73144.1 KQ979685 KYN19837.1
PCG64991.1 ODYU01005101 SOQ45643.1 GAIX01004400 JAA88160.1 AGBW02011959 OWR45868.1 JTDY01004323 KOB68296.1 GFDF01003523 JAV10561.1 GDAI01000035 JAI17568.1 AJVK01021645 DS231904 EDS45209.1 GFDL01012596 JAV22449.1 ATLV01023433 KE525343 KFB49166.1 AXCM01005445 APCN01001695 GGFJ01005376 MBW54517.1 AAAB01008980 EAA14036.3 NEVH01016304 PNF25753.1 GDHF01016458 GDHF01010834 GDHF01009540 GDHF01004598 JAI35856.1 JAI41480.1 JAI42774.1 JAI47716.1 ADMH02001962 ETN60222.1 GAMC01002909 JAC03647.1 CH477933 EAT34957.1 GAKP01008961 GAKP01008960 GAKP01008959 JAC49993.1 AXCN02000654 GEZM01080460 JAV62131.1 GBXI01014394 JAC99897.1 UFQT01001122 SSX29027.1 GANO01000552 JAB59319.1 CH954177 EDV59038.1 CH480820 EDW54475.1 CM000361 CM002910 EDX03917.1 KMY88466.1 UFQT01003842 SSX35305.1 CH933807 EDW11745.1 CM000157 EDW87935.1 AE014134 BT044528 AAF52344.1 ACH95302.1 AY060647 AAL28195.1 CH902620 EDV31531.1 CH379058 EAL34247.1 OUUW01000010 SPP86121.1 CH940649 EDW63623.1 GGFK01008910 MBW42231.1 CH479180 EDW28204.1 CH916368 EDW02870.1 JRES01001249 KNC24246.1 CP012523 ALC38368.1 ALC38182.1 CVRI01000047 CRK97785.1 GEDC01003327 JAS33971.1 JXUM01126916 JXUM01126917 JXUM01126918 KQ567125 KXJ69610.1 GBGD01001545 JAC87344.1 CH963857 EDW76306.1 GECL01003599 JAP02525.1 GFTR01006154 JAW10272.1 KK852432 KDR23972.1 JXJN01016549 CCAG010006198 PNF25754.1 APGK01006704 APGK01034104 KB740904 KB736898 KB632006 ENN78434.1 ENN83113.1 ERL87945.1 ACPB03022604 GAHY01000468 JAA77042.1 GDKW01000388 JAI56207.1 KQ760343 OAD60859.1 KQ414725 KOC62665.1 GBHO01016970 GBRD01001301 GDHC01016797 GDHC01001501 JAG26634.1 JAG64520.1 JAQ01832.1 JAQ17128.1 KQ971343 EFA04425.1 KZ288215 PBC32769.1 KK107109 QOIP01000008 EZA59226.1 RLU19695.1 GEBQ01017339 JAT22638.1 GEDC01009403 JAS27895.1 GL888186 EGI65515.1 GEMB01002673 JAS00520.1 LBMM01001611 KMQ96015.1 PYGN01000035 PSN56741.1 ADTU01021228 KQ981208 KYN44714.1 KQ435803 KOX73144.1 KQ979685 KYN19837.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000092462
UP000002320
+ More
UP000075880 UP000030765 UP000075881 UP000075883 UP000075840 UP000075884 UP000007062 UP000075903 UP000075882 UP000075902 UP000235965 UP000075885 UP000075901 UP000076407 UP000076408 UP000069272 UP000000673 UP000075900 UP000075920 UP000008820 UP000075886 UP000095301 UP000008711 UP000001292 UP000000304 UP000095300 UP000192221 UP000009192 UP000002282 UP000000803 UP000007801 UP000001819 UP000268350 UP000008792 UP000008744 UP000001070 UP000037069 UP000092553 UP000183832 UP000069940 UP000249989 UP000007798 UP000027135 UP000078200 UP000192223 UP000092445 UP000092460 UP000092443 UP000092444 UP000019118 UP000030742 UP000015103 UP000053825 UP000007266 UP000242457 UP000053097 UP000279307 UP000005203 UP000007755 UP000036403 UP000245037 UP000005205 UP000078541 UP000053105 UP000078492
UP000075880 UP000030765 UP000075881 UP000075883 UP000075840 UP000075884 UP000007062 UP000075903 UP000075882 UP000075902 UP000235965 UP000075885 UP000075901 UP000076407 UP000076408 UP000069272 UP000000673 UP000075900 UP000075920 UP000008820 UP000075886 UP000095301 UP000008711 UP000001292 UP000000304 UP000095300 UP000192221 UP000009192 UP000002282 UP000000803 UP000007801 UP000001819 UP000268350 UP000008792 UP000008744 UP000001070 UP000037069 UP000092553 UP000183832 UP000069940 UP000249989 UP000007798 UP000027135 UP000078200 UP000192223 UP000092445 UP000092460 UP000092443 UP000092444 UP000019118 UP000030742 UP000015103 UP000053825 UP000007266 UP000242457 UP000053097 UP000279307 UP000005203 UP000007755 UP000036403 UP000245037 UP000005205 UP000078541 UP000053105 UP000078492
Interpro
ProteinModelPortal
H9J9B6
Q1HQ78
A0A2W1BGI8
A0A2A4IZ04
A0A2H1VXU2
S4PKN2
+ More
A0A212EWJ1 A0A0L7KZ05 A0A1L8DVU4 A0A0K8TTU7 A0A1B0D0S7 B0WE33 A0A1Q3F4K2 A0A182JN74 A0A084WG22 A0A182K7U2 A0A182MR26 A0A182HRE2 A0A182NAV9 A0A2M4BN61 Q7Q168 A0A182V0D8 A0A182KYZ7 A0A182UFT3 A0A2J7QAZ4 A0A182PG50 A0A0K8VSB4 A0A182SHZ7 A0A182XHT4 A0A182Y6E5 A0A182F4Q4 W5J6Y9 W8C8M4 A0A182RRB9 A0A182W8X1 Q16KX6 A0A034W887 A0A182QV26 A0A1Y1KNU1 A0A0A1WNB4 A0A336MKY6 U5EWI6 A0A1I8MZN2 B3N5F9 B4I1R8 B4Q4J0 A0A336N041 A0A1I8QAQ8 A0A1W4VAT6 B4KL84 B4P0P0 Q9VMH4 Q95SQ7 B3MKH8 Q29PP5 A0A3B0KM02 B4LRM8 A0A2M4AN82 B4G6H9 B4JB00 A0A0L0BW54 A0A0M5J8U9 A0A0M4ENX0 A0A1J1IH73 A0A1B6E7Q3 A0A182HC93 A0A069DYZ2 B4MUW1 A0A0V0G349 A0A224XNX5 A0A067RJH6 A0A1A9V1A3 A0A1W4WWP5 A0A1B0A875 A0A1B0BLT1 A0A1A9Y7J3 A0A1B0G0U4 A0A2J7QB04 N6TDI8 R4FNS0 A0A0P4VU66 A0A310SU50 A0A0L7QVM4 A0A0A9Y3A6 D6WP28 A0A2A3ENR3 A0A026WU94 A0A1B6LG88 A0A1B6DQD1 A0A088A1C2 F4WJM3 A0A170Z2L7 A0A0J7NUD4 A0A2P8ZJR5 A0A158NNC0 A0A195FY01 A0A0N0U4Q0 A0A195E4K8
A0A212EWJ1 A0A0L7KZ05 A0A1L8DVU4 A0A0K8TTU7 A0A1B0D0S7 B0WE33 A0A1Q3F4K2 A0A182JN74 A0A084WG22 A0A182K7U2 A0A182MR26 A0A182HRE2 A0A182NAV9 A0A2M4BN61 Q7Q168 A0A182V0D8 A0A182KYZ7 A0A182UFT3 A0A2J7QAZ4 A0A182PG50 A0A0K8VSB4 A0A182SHZ7 A0A182XHT4 A0A182Y6E5 A0A182F4Q4 W5J6Y9 W8C8M4 A0A182RRB9 A0A182W8X1 Q16KX6 A0A034W887 A0A182QV26 A0A1Y1KNU1 A0A0A1WNB4 A0A336MKY6 U5EWI6 A0A1I8MZN2 B3N5F9 B4I1R8 B4Q4J0 A0A336N041 A0A1I8QAQ8 A0A1W4VAT6 B4KL84 B4P0P0 Q9VMH4 Q95SQ7 B3MKH8 Q29PP5 A0A3B0KM02 B4LRM8 A0A2M4AN82 B4G6H9 B4JB00 A0A0L0BW54 A0A0M5J8U9 A0A0M4ENX0 A0A1J1IH73 A0A1B6E7Q3 A0A182HC93 A0A069DYZ2 B4MUW1 A0A0V0G349 A0A224XNX5 A0A067RJH6 A0A1A9V1A3 A0A1W4WWP5 A0A1B0A875 A0A1B0BLT1 A0A1A9Y7J3 A0A1B0G0U4 A0A2J7QB04 N6TDI8 R4FNS0 A0A0P4VU66 A0A310SU50 A0A0L7QVM4 A0A0A9Y3A6 D6WP28 A0A2A3ENR3 A0A026WU94 A0A1B6LG88 A0A1B6DQD1 A0A088A1C2 F4WJM3 A0A170Z2L7 A0A0J7NUD4 A0A2P8ZJR5 A0A158NNC0 A0A195FY01 A0A0N0U4Q0 A0A195E4K8
PDB
5TF2
E-value=7.50131e-49,
Score=490
Ontologies
GO
Topology
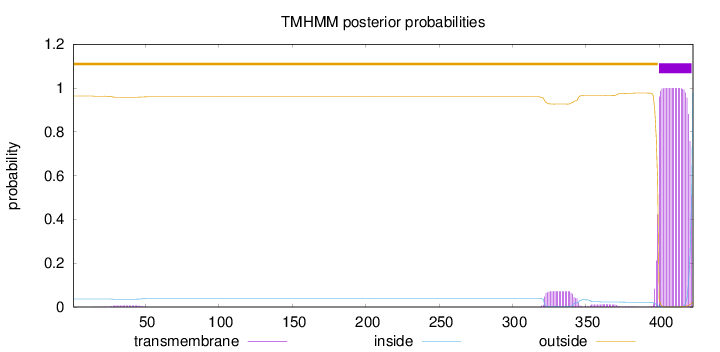
Length:
423
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.68873
Exp number, first 60 AAs:
0.13569
Total prob of N-in:
0.03680
outside
1 - 399
TMhelix
400 - 422
inside
423 - 423
Population Genetic Test Statistics
Pi
202.72022
Theta
178.876053
Tajima's D
-0.807471
CLR
0.88249
CSRT
0.179391030448478
Interpretation
Uncertain
