Gene
KWMTBOMO01668
Pre Gene Modal
BGIBMGA006106
Annotation
PREDICTED:_uncharacterized_protein_LOC106136881_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.862
Sequence
CDS
ATGGGCGAGGTACCGTGCGACTGGCAGTTGGCCTGCACCGAAATTAAACAACGAGGAGAGTATTTGCTTCAATCTGAACAATGGTCAGATTGTACCTTCCTGGTGGGATCTGAGCCGAACCAAGTTGAGGTGCGTGGCCACAGGCTCATCCTGGCAATGGCTTCTCCTGTTTTCGAAGCTATGTTCTACGGTGGAATGGCTGAGAGAAATGATCCGATCCCTATTCTAGATGTCCAGCCTGAAGCATTCAAAGCGCTTTTAGAATACATTTACACGGATAACATCAACATCAGTTCATTCGATAAAGCATGTGAATTGTGCTATGGCGCCAAAAAGTACATGCTTCCGCATCTAGTCAAGGAGTGCACTAAGTATCTGTGGTCCGATCTGTACCCTCGCAATGCGTGCCGAGCCTATGAATTCGCCCGTCTCTTCGAAGAGAGCGTGCTGATGGACAAATGTCTCCAGATTATCAGCACTAACACCAAAGAGGTGCTCCACGACTGTAGCTTTGAGGAAGTGGAGCTCAACACGCTGATCACTGTGTTCTCCTTGGAACACTTGAACATCGACAGTGAGCTGGATCTGTTCGAAGCTGCTGTTAGATATGCCAAGGGACAGGAGAGGAGGAAGGACGACCGGAGCGCTAGCCCGCCTGCTGATGGACCGCCAGCCGAGAAAAGGTCCAAGAGTCCTCAGCCGTGTACTTCACAGGAGAAGGTGGTAAACGTAGAGAGCAGCGCTGAGAACAGTCCTGCCGCTACAACCGATGCTACGAAGCCAGATGAAGAACCGGAAACAACTCCAGTTCCTGAGGAGACCAAGAAGGAAAAGAGAGTTGAGCCTAAGGACAACAAGCCCACCATCCGCAAGGCGATCGAGAAAATTCGCTTCCTGACGCTGACTCCTCAGCAGTTCGCCAAGGGTCCGGCCAAGTCCTCCCTGCTCACCGAGTCTGAAGCATTTGCAGTCCTCATGAACATCTTGTCCAGCAATGCTGATGTGCCCATGCCCAGAGGCTTTTCTACTTGCAGAGTTCCCAGGAAGCAACTCATCGGTGGTCCCCAGTCCTCCAATATCCCGACGCTGACCGTGGACACCCCGAGCCCCGTGATGCTGGAGCAGGTGTTCGGCAACGAGTCCTCGCGCGGCCACTCGCTCAACGTGTCGCACCCGCACCACCACCACTGCGTCATGCAGTTCCCCAGGCACCCCAGGCTCGAAGGTGTACGTATGTCATCAATGATGAACAATCCGGTGTTGAACGGTATCGGCGTGGAGCACATGGAGCGCGGGGCTGAGATCAGCAAGATCTACTGCCAGCGCCCCATCATCCAGCACACCGACTGCCTCAACACCAGCGTCCTCGACTGCTCCGTCACCTTCATGGTCGACAAGAATATCTGCCTCCTCGGTGTCCAGGTGCCGACGCAAGCTCCGAGCGAGGAAGGCGCTGCGAGCGCTGTGCCGGGCGTGGCGGGCTACTCGGAGCTGCTGTACGCGCACCTGCTGGACGCGGACGGAGCGCGCCTCACCTACACGCACTACACCAGCCGCGTACCCTACCGACACCTGCTCGACATCATGTTCAACCGACCCGTGTACATCCAGAGGAACAAGGTGTACAAGGTGGGGATAGTGTTCAACAAAGTGGGCTGGTACCCGATGGGCACGTGCGCGCAGCAGGTGGCCGTCGAGACGGTCTTCTTCAACTTCGGAATTGGGCAGATCAATGATTCGGTCCGCGACGGTCTCATCCGCTCCATCATCTTCACGTACTAG
Protein
MGEVPCDWQLACTEIKQRGEYLLQSEQWSDCTFLVGSEPNQVEVRGHRLILAMASPVFEAMFYGGMAERNDPIPILDVQPEAFKALLEYIYTDNINISSFDKACELCYGAKKYMLPHLVKECTKYLWSDLYPRNACRAYEFARLFEESVLMDKCLQIISTNTKEVLHDCSFEEVELNTLITVFSLEHLNIDSELDLFEAAVRYAKGQERRKDDRSASPPADGPPAEKRSKSPQPCTSQEKVVNVESSAENSPAATTDATKPDEEPETTPVPEETKKEKRVEPKDNKPTIRKAIEKIRFLTLTPQQFAKGPAKSSLLTESEAFAVLMNILSSNADVPMPRGFSTCRVPRKQLIGGPQSSNIPTLTVDTPSPVMLEQVFGNESSRGHSLNVSHPHHHHCVMQFPRHPRLEGVRMSSMMNNPVLNGIGVEHMERGAEISKIYCQRPIIQHTDCLNTSVLDCSVTFMVDKNICLLGVQVPTQAPSEEGAASAVPGVAGYSELLYAHLLDADGARLTYTHYTSRVPYRHLLDIMFNRPVYIQRNKVYKVGIVFNKVGWYPMGTCAQQVAVETVFFNFGIGQINDSVRDGLIRSIIFTY
Summary
Uniprot
A0A2W1BM81
A0A2H1VYP0
A0A194Q1P5
A0A212FBQ3
A0A2A4J0Q3
A0A0L7KWB3
+ More
A0A1W4WSP4 A0A0T6BCN3 D6WNZ2 A0A023EV13 A0A1S4G607 J9HTU8 B0W0L6 A0A1Q3FDJ8 A0A1B0DD34 J3JU42 A0A1A9VBM9 A0A1J1J3N0 A0A336LFN0 A0A1B0G8D1 A0A1B0C7L6 A0A1A9Z5U1 A0A164RRV5 A0A0P5XYQ9 A0A0P5DLQ1 A0A0P6BIG9 A0A0P6HWD7 A0A0P5B8H3 A0A0P5S8E5
A0A1W4WSP4 A0A0T6BCN3 D6WNZ2 A0A023EV13 A0A1S4G607 J9HTU8 B0W0L6 A0A1Q3FDJ8 A0A1B0DD34 J3JU42 A0A1A9VBM9 A0A1J1J3N0 A0A336LFN0 A0A1B0G8D1 A0A1B0C7L6 A0A1A9Z5U1 A0A164RRV5 A0A0P5XYQ9 A0A0P5DLQ1 A0A0P6BIG9 A0A0P6HWD7 A0A0P5B8H3 A0A0P5S8E5
Pubmed
EMBL
KZ150155
PZC72793.1
ODYU01005101
SOQ45642.1
KQ459579
KPI99471.1
+ More
AGBW02009291 OWR51174.1 NWSH01004520 PCG64992.1 JTDY01005069 KOB67405.1 LJIG01002079 KRT84861.1 KQ971343 EFA04408.1 JXUM01047493 GAPW01000655 KQ561521 JAC12943.1 KXJ78297.1 CH478175 EJY58078.1 DS231818 EDS41318.1 GFDL01009400 JAV25645.1 AJVK01031598 BT126754 AEE61716.1 CVRI01000069 CRL06976.1 UFQS01004266 UFQT01004266 SSX16294.1 SSX35614.1 CCAG010000240 CCAG010000241 CCAG010000242 JXJN01029041 LRGB01002167 KZS08886.1 GDIP01078354 JAM25361.1 GDIP01155398 JAJ68004.1 GDIP01014577 JAM89138.1 GDIQ01021142 JAN73595.1 GDIP01192384 JAJ31018.1 GDIQ01091085 JAL60641.1
AGBW02009291 OWR51174.1 NWSH01004520 PCG64992.1 JTDY01005069 KOB67405.1 LJIG01002079 KRT84861.1 KQ971343 EFA04408.1 JXUM01047493 GAPW01000655 KQ561521 JAC12943.1 KXJ78297.1 CH478175 EJY58078.1 DS231818 EDS41318.1 GFDL01009400 JAV25645.1 AJVK01031598 BT126754 AEE61716.1 CVRI01000069 CRL06976.1 UFQS01004266 UFQT01004266 SSX16294.1 SSX35614.1 CCAG010000240 CCAG010000241 CCAG010000242 JXJN01029041 LRGB01002167 KZS08886.1 GDIP01078354 JAM25361.1 GDIP01155398 JAJ68004.1 GDIP01014577 JAM89138.1 GDIQ01021142 JAN73595.1 GDIP01192384 JAJ31018.1 GDIQ01091085 JAL60641.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BM81
A0A2H1VYP0
A0A194Q1P5
A0A212FBQ3
A0A2A4J0Q3
A0A0L7KWB3
+ More
A0A1W4WSP4 A0A0T6BCN3 D6WNZ2 A0A023EV13 A0A1S4G607 J9HTU8 B0W0L6 A0A1Q3FDJ8 A0A1B0DD34 J3JU42 A0A1A9VBM9 A0A1J1J3N0 A0A336LFN0 A0A1B0G8D1 A0A1B0C7L6 A0A1A9Z5U1 A0A164RRV5 A0A0P5XYQ9 A0A0P5DLQ1 A0A0P6BIG9 A0A0P6HWD7 A0A0P5B8H3 A0A0P5S8E5
A0A1W4WSP4 A0A0T6BCN3 D6WNZ2 A0A023EV13 A0A1S4G607 J9HTU8 B0W0L6 A0A1Q3FDJ8 A0A1B0DD34 J3JU42 A0A1A9VBM9 A0A1J1J3N0 A0A336LFN0 A0A1B0G8D1 A0A1B0C7L6 A0A1A9Z5U1 A0A164RRV5 A0A0P5XYQ9 A0A0P5DLQ1 A0A0P6BIG9 A0A0P6HWD7 A0A0P5B8H3 A0A0P5S8E5
PDB
2VKP
E-value=7.01306e-16,
Score=207
Ontologies
GO
Topology
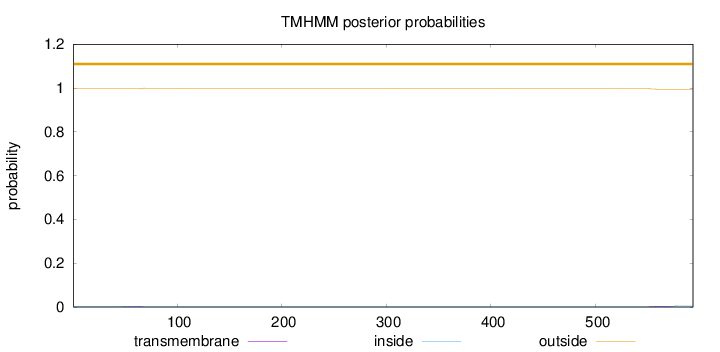
Length:
593
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17032
Exp number, first 60 AAs:
0.04277
Total prob of N-in:
0.00373
outside
1 - 593
Population Genetic Test Statistics
Pi
274.413244
Theta
181.884933
Tajima's D
1.319437
CLR
0.609451
CSRT
0.744112794360282
Interpretation
Uncertain
