Pre Gene Modal
BGIBMGA006044
Annotation
PREDICTED:_pre-mRNA-splicing_factor_CWC22_homolog_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.289
Sequence
CDS
ATGGAGAAGACTAACACCAAGCAGAAGAAAGATAAATACGAGGAAGATGATGAAAGGCGTAGGTCGAAAAGGTCGAGACGGGCTAGATCTAGATCTAAGTCACTTGAGGAACAGCAGAATAGGGAGTATAGAAGTAAACGCCGGCGTTCCAAATCACGCTCACCTCATTACAAGGCTAGGGAAGATAAACGAGTTAAAGACACACAAAAAGAGCCAGAAACAAAAAAAGATGCCTCTACAAAGCCAGAAAGACGCGCAAAAGATACTGATATGTTGAATACTAGAACAGGAGGTGCATACATTCCACCAGCAAGACTGAGGATGATGCAAGCGCAGATCACGGATAAGTCCTCGATAGCTTACCAAAGACTTGCCTGGGAAGCTCTCAAAAAGTCAATTCACGGTCACATCAACAAAATTAATGTCGGTAACATAGGTATCATTATAAAGGAATTATTGAAAGAAAACATTGTCAGAGGTCGAGGTTTGTTATGCCGTTCTGTTATTCAAGCCCAGGCAGCCTCGCCAACATTCACTAATGTATACGCGGCTCTAGTATCAGCAGTCAATTCACGTTTTCCTAATATCGGGGAGTTGTTACTGAAAAGATTAGTGATTCAATTCAAAAGAGGTTTTAAACGCAATGATAAGTCAATTTGCATTTCCTCTGCATCTTTCATTGCACATTTAGTGAATCAGAGAGTGGCACATGAAATTCTGGCTTTAGAACTTCTCACTTTACTAGTTGAATCACCAACAGATGATTCAGTTGAAGTTGCCATAGCATTCTTAAAGGAATGTGGCCAAAAATTGACTGAAGTATCATCTAAAGGTGTAAATGCAATTTTCGAGATGCTAAGGAACATTTTGCACGAAGGTCAGTTAGACAAAAGAGTTCAGTACATGATAGAAGTAATGTTCCAAGTTTGGAAGGATGGGTTCAAAGACCACCCGGCACTGATTGAAGAGTTGGAGCTCGTGCCTGAAGAAGAACAGTTCACACATCTATTGATGCTGGATGACGCTACAGATCCTCAAGATATCCTGAATGTTTTCAAATTTGATGAAAAGTATGAAGAGAATGAACAGAAGTACAAGAATCTTTGCAAGGAAATACTTGGTTCTGATGCTGAGTCCGGTGATGAAGATGGTTCAGGAGAAAGCGGTAGTGAGGAATCTGATGAAGAAGATGAAGAACAACCAAAAAAAGAAATGACTATTATTGATAACACAGAAACAAACTTAGTGGCTCTGAGACGCACTATATACCTGACAATCAACTCCAGTTTAGATTTCGAAGAATGTGCTCATAAACTAATGAAGATGCAACTGAAACCAGGACAAGAAGTGGAACTATGCCACATGTTCTTAGACTGCTGTGCTGAGCAGAGAACATATGAGAAATTCTATGGCTTATTGGCCCAAAGGTTTTGCAACATCAACAGAATATACATCGGACCTTTCGAGGAAATTTTCAAAGATTCATATTCAACAGCACACAGGTTGGACACGAATAGACTCAGAAATGTCAGCAAGTTTTTCGCCCATCTGCTGTTTACAGATTCCATTAGTTGGGAATCTTTGGAATGTGTTAAATTGAACGAAGAAGACACAACTAGTTCTAGTCGGATATACATAAAGATTCTGTTTCAAGAACTGGCTGAATACATGGGACTAAAGAAACTGAATGACCGTCTACAAGATCCGACGTTACAAGAGGCGTTTGCCGGACTGTTTCCCCGCGACAACCCGAGAAACACGCGGTTCTCTATAAACTTCTTCACGTCCATCGGACTTGGTGGACTAACTGATGAGTTACGTGAGCACCTCAAACAAATGCCCAAGAATACACCGGCGCCACCAACAGAGATCAAAATAGACAGTGACTCCAGCTCTGACTCCAGCTCGGATTCGTCTTCTTCAAGTGATTCCAGCTCAAGCGAATCTGAGGAAGAAACACAGAAAAAGAAGAAAGAAAAGAATAAAAACAAGGAAAAATCACCAGAACCAGAAAAACCCCAAGACAATAAGGTCGAGCGGTGGGGACATGAGGGGTTCTACGATACTTACGGGAGGGACGCGCGGCGACACGACCCTCGCGACCGCGGCGACGGGGGGCGGGGGGGCGCACACCGGGATACAGACCAACAAGGAAGACGTGCCAGGGACGAGAACGGGAGAGGTCTGAGAAGACGAGATGAAGATCACGAGCAAGACCCAAGAGATAGAAGAGCTCCAGAAAAGGATGACAGGAGGAATCGAAAGGAGGGTGAAAAGAGTCGTGATGGGAACAGTCGTAAAGATATTGATAGAGACAGTGAGAAAGAAAAGGATAAGAGAAGTCGAAAAGATGATGACAGAGTAACTGACAGGGAAAAGGATAAGAGACAGACCAGGAACAGTGAAGAAGATAGTAGAAGACGGGACTCTGAATACAATAGAAAAGGTGAAAAAGAGAAGGAAAAGAAACGAAAGAACGCCCGGGACGATGAACACAAAAAGGAAACATATCACAATGATGAATATTCAGCTCGAGACGAGACCGAGTCCAGGAGAACTAAGGACAAGAGAAGTAGGAATGCGGGCAGTGACGACGACAGGAAACAGAGAGGGCACAGTAGTGACGAGGGCCGCCGCGAGCGTCGCACTAGGAAAGACAAACAAAAGAGAGGCGGCAGGAGGAAGAGTGATGCTTCTGACGACGACGACGGCTGGGATATCAAATGCAGGCAGAATGCACCGAATGAGGTCTCCGGGAGCAAGTGCTATGCCGCGATGCTCGATTCTAAAAGTAACTCTGGGTCAGTGAGCTCGGCGGCTCTCAGGGACAAGGCCGCGTCGGATCCGTCTTGTGCCCCTCAGAATGTAACCGAGGCTCAGATAAAGCCACAGATCTCTGCACCCTCGTATTTTTATTCTTTGAAGGAGAACGAAGCGTTGCCGTTCAAGAGGCCCGAGAAAGAACCGCGACCAAAACTACTGCTGAATAGCGTCGTGAACGAAAACGATGTTGTGGCGATGATCGCCGCGCATGACGCGAACAGAACTGAAGGTCTGAAGAAACACATAGGTAAACTTTTAGTCGAAAAGCAAATCGTGGTCAATTATAATTTGTCAGAAAGAAAATTTGTCACGAATCTGCTGTGCGAGACGATAGACTATGCAGCCCAGCGCGACTTCTCGGCGTTTAAGCTAGCTTGTTTACTTACAACCTACGTCACTGTTCATAGTTACTTCAAGTGGTATTATTGGCAAGCCCCAGTCTCTGTTTGGATGTATTTCAAAGAACTTATGATACGGCTTACAATTGAGGTGATGAACAAATTTATTTTCAAACACTTTCTTTTCGAAGTCATATTTTCTTGA
Protein
MEKTNTKQKKDKYEEDDERRRSKRSRRARSRSKSLEEQQNREYRSKRRRSKSRSPHYKAREDKRVKDTQKEPETKKDASTKPERRAKDTDMLNTRTGGAYIPPARLRMMQAQITDKSSIAYQRLAWEALKKSIHGHINKINVGNIGIIIKELLKENIVRGRGLLCRSVIQAQAASPTFTNVYAALVSAVNSRFPNIGELLLKRLVIQFKRGFKRNDKSICISSASFIAHLVNQRVAHEILALELLTLLVESPTDDSVEVAIAFLKECGQKLTEVSSKGVNAIFEMLRNILHEGQLDKRVQYMIEVMFQVWKDGFKDHPALIEELELVPEEEQFTHLLMLDDATDPQDILNVFKFDEKYEENEQKYKNLCKEILGSDAESGDEDGSGESGSEESDEEDEEQPKKEMTIIDNTETNLVALRRTIYLTINSSLDFEECAHKLMKMQLKPGQEVELCHMFLDCCAEQRTYEKFYGLLAQRFCNINRIYIGPFEEIFKDSYSTAHRLDTNRLRNVSKFFAHLLFTDSISWESLECVKLNEEDTTSSSRIYIKILFQELAEYMGLKKLNDRLQDPTLQEAFAGLFPRDNPRNTRFSINFFTSIGLGGLTDELREHLKQMPKNTPAPPTEIKIDSDSSSDSSSDSSSSSDSSSSESEEETQKKKKEKNKNKEKSPEPEKPQDNKVERWGHEGFYDTYGRDARRHDPRDRGDGGRGGAHRDTDQQGRRARDENGRGLRRRDEDHEQDPRDRRAPEKDDRRNRKEGEKSRDGNSRKDIDRDSEKEKDKRSRKDDDRVTDREKDKRQTRNSEEDSRRRDSEYNRKGEKEKEKKRKNARDDEHKKETYHNDEYSARDETESRRTKDKRSRNAGSDDDRKQRGHSSDEGRRERRTRKDKQKRGGRRKSDASDDDDGWDIKCRQNAPNEVSGSKCYAAMLDSKSNSGSVSSAALRDKAASDPSCAPQNVTEAQIKPQISAPSYFYSLKENEALPFKRPEKEPRPKLLLNSVVNENDVVAMIAAHDANRTEGLKKHIGKLLVEKQIVVNYNLSERKFVTNLLCETIDYAAQRDFSAFKLACLLTTYVTVHSYFKWYYWQAPVSVWMYFKELMIRLTIEVMNKFIFKHFLFEVIFS
Summary
Uniprot
Pubmed
Proteomes
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
PDB
6QDV
E-value=7.11063e-170,
Score=1537
Ontologies
KEGG
GO
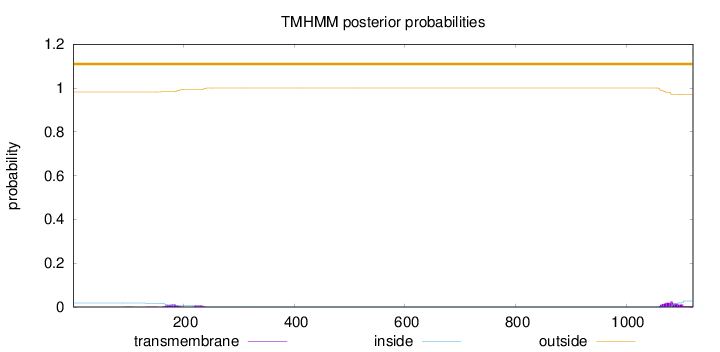
Topology
Length:
1121
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.06617
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01794
outside
1 - 1121
Population Genetic Test Statistics
Pi
227.688868
Theta
187.355734
Tajima's D
0.620407
CLR
0.328581
CSRT
0.550672466376681
Interpretation
Uncertain
