Pre Gene Modal
BGIBMGA006105
Annotation
PREDICTED:_anaphase-promoting_complex_subunit_13-like_isoform_X2_[Bombyx_mori]
Full name
Anaphase-promoting complex subunit 13
Alternative Name
Cyclosome subunit 13
Location in the cell
Nuclear Reliability : 2.713
Sequence
CDS
ATGGACAGTAAGTGTCGCCGTAACGGTCGCCTAATAGACATAGTTGATGAGGACTGGCGTGCGGAACAACTTCCTCACGAGGACATTCAAGTTCCGCTGGATGAGCTCCCGGACCCCGAGGCAGACAGCGCCGACTCTCATCTCACTCTAAAAGAGCAGAGCATGCGTTGGCAGGAGAATTTCCCTGAACCAGCGAGCAGTAATCAAAACTAG
Protein
MDSKCRRNGRLIDIVDEDWRAEQLPHEDIQVPLDELPDPEADSADSHLTLKEQSMRWQENFPEPASSNQN
Summary
Description
Component of the anaphase promoting complex/cyclosome (APC/C), a cell cycle-regulated E3 ubiquitin ligase that controls progression through mitosis and the G1 phase of the cell cycle. The APC/C complex acts by mediating ubiquitination and subsequent degradation of target proteins: it mainly mediates the formation of 'Lys-11'-linked polyubiquitin chains and, to a lower extent, the formation of 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains (By similarity).
Subunit
The mammalian APC/C is composed at least of 14 distinct subunits ANAPC1, ANAPC2, CDC27/APC3, ANAPC4, ANAPC5, CDC16/APC6, ANAPC7, CDC23/APC8, ANAPC10, ANAPC11, CDC26/APC12, ANAPC13, ANAPC15 and ANAPC16 that assemble into a complex of at least 19 chains with a combined molecular mass of around 1.2 MDa; APC/C interacts with FZR1 and FBXO5.
Similarity
Belongs to the APC13 family.
Keywords
Cell cycle
Cell division
Complete proteome
Mitosis
Nucleus
Reference proteome
Ubl conjugation pathway
Feature
chain Anaphase-promoting complex subunit 13
Uniprot
H9J9B3
S4PHD2
A0A212EGU5
A0A194PZN4
A0A0L7KXH9
A0A0L7KUM8
+ More
A0A2A3EQZ0 A0A088AF80 A0A224Y328 A0A069DNR7 A0A023FA82 A0A0V0GDT8 A0A2P8YVU8 A0A154P1T5 A0A067QVD4 A0A0P4VP53 R4G309 A0A139WH34 A0A310S9R7 E2BKZ3 A0A2M4AY03 A0A151IKA6 A0A195EEQ7 F4W9W8 A0A195EY46 A0A158NZI5 A0A151WKR5 A0A182XEV2 A0A182UK85 A0A182HNX8 A0A2M3ZCW0 W5JRS3 A0A0J7KQC2 E2A4Q6 Q7PQ30 A0A2M4C550 A0A026WAL0 A0A0L7QYQ3 A0A1I8PIM1 A0A182QYI4 A0A182MLP5 A0A182RKW0 A0A0K8R6K0 B7Q8J4 A0A2J7QCQ4 A0A182P8S8 A0A182K7W3 K7JCE8 A0A1I8N725 A0A336LCX2 A0A182G703 Q5RBV4 A0A2L2YHC5 A0A182WK22 A0A2J7RD63 U4UPW5 A0A232EDF1 A0A087U180 D3TRJ1 A0A0C9RZU1 A0A1B0FP98 H3AY45 N6TUI7 A0A226NCN8 A0A1L8DCU8 A0A091KF47 B5G2E5 A0A0A0ANS5 A0A093I9H5 A0A093CP24 A0A093ERQ3 A0A091HUA8 A0A091UW42 A0A091WL26 A0A087V753 A0A091LLC4 A0A091MLG3 A0A093DNA7 A0A091KA51 A0A091TG81 A0A2I0LUR1 A0A093R7P2 A0A093GNC1 A0A094KGE7 A0A093J3S4 A0A093H1N3 A0A091QD37 A0A087RD33 R0JW70 B5G2E3 A0A1U7RN24 A0A093R9D4 A0A091EI49 G1QBV4 A0A1S2ZMX8 S7PDV3 A0A1D5PVV7 I3N837 G3SST0 U6CR27
A0A2A3EQZ0 A0A088AF80 A0A224Y328 A0A069DNR7 A0A023FA82 A0A0V0GDT8 A0A2P8YVU8 A0A154P1T5 A0A067QVD4 A0A0P4VP53 R4G309 A0A139WH34 A0A310S9R7 E2BKZ3 A0A2M4AY03 A0A151IKA6 A0A195EEQ7 F4W9W8 A0A195EY46 A0A158NZI5 A0A151WKR5 A0A182XEV2 A0A182UK85 A0A182HNX8 A0A2M3ZCW0 W5JRS3 A0A0J7KQC2 E2A4Q6 Q7PQ30 A0A2M4C550 A0A026WAL0 A0A0L7QYQ3 A0A1I8PIM1 A0A182QYI4 A0A182MLP5 A0A182RKW0 A0A0K8R6K0 B7Q8J4 A0A2J7QCQ4 A0A182P8S8 A0A182K7W3 K7JCE8 A0A1I8N725 A0A336LCX2 A0A182G703 Q5RBV4 A0A2L2YHC5 A0A182WK22 A0A2J7RD63 U4UPW5 A0A232EDF1 A0A087U180 D3TRJ1 A0A0C9RZU1 A0A1B0FP98 H3AY45 N6TUI7 A0A226NCN8 A0A1L8DCU8 A0A091KF47 B5G2E5 A0A0A0ANS5 A0A093I9H5 A0A093CP24 A0A093ERQ3 A0A091HUA8 A0A091UW42 A0A091WL26 A0A087V753 A0A091LLC4 A0A091MLG3 A0A093DNA7 A0A091KA51 A0A091TG81 A0A2I0LUR1 A0A093R7P2 A0A093GNC1 A0A094KGE7 A0A093J3S4 A0A093H1N3 A0A091QD37 A0A087RD33 R0JW70 B5G2E3 A0A1U7RN24 A0A093R9D4 A0A091EI49 G1QBV4 A0A1S2ZMX8 S7PDV3 A0A1D5PVV7 I3N837 G3SST0 U6CR27
Pubmed
19121390
23622113
22118469
26354079
26227816
26334808
+ More
25474469 29403074 24845553 27129103 18362917 19820115 20798317 21719571 21347285 20920257 23761445 12364791 24508170 30249741 20075255 25315136 26483478 26561354 23537049 28648823 20353571 26131772 9215903 17018643 23371554 21993624 15592404
25474469 29403074 24845553 27129103 18362917 19820115 20798317 21719571 21347285 20920257 23761445 12364791 24508170 30249741 20075255 25315136 26483478 26561354 23537049 28648823 20353571 26131772 9215903 17018643 23371554 21993624 15592404
EMBL
BABH01004417
GAIX01000509
JAA92051.1
AGBW02015019
OWR40704.1
KQ459584
+ More
KPI98498.1 JTDY01004718 KOB67840.1 JTDY01005705 KOB66719.1 KZ288199 PBC33636.1 GFTR01000906 JAW15520.1 GBGD01003657 JAC85232.1 GBBI01000798 JAC17914.1 GECL01000260 JAP05864.1 PYGN01000332 PSN48374.1 KQ434803 KZC05909.1 KK852976 KDR13050.1 GDKW01002472 JAI54123.1 ACPB03020263 GAHY01002106 JAA75404.1 KQ971343 KYB27308.1 KQ767877 OAD53281.1 GL448858 EFN83647.1 GGFK01012352 MBW45673.1 KQ977234 KYN04703.1 KQ979039 KYN23334.1 GL888033 EGI69104.1 KQ981909 KYN33205.1 ADTU01000887 KQ983012 KYQ48371.1 APCN01002811 GGFM01005636 MBW26387.1 ADMH02000362 ETN66831.1 LBMM01004382 KMQ92481.1 GL436716 EFN71590.1 AAAB01008900 EAA09386.5 GGFJ01011289 MBW60430.1 KK107321 QOIP01000002 EZA52696.1 RLU25393.1 KQ414688 KOC63708.1 AXCN02001020 AXCM01001015 GADI01007021 JAA66787.1 ABJB010972031 DS884446 EEC15166.1 NEVH01016057 PNF26369.1 AAZX01003377 UFQS01002630 UFQT01002630 SSX14405.1 SSX33815.1 JXUM01149326 KQ570750 KXJ68307.1 CR858529 IAAA01022778 LAA07509.1 NEVH01005296 PNF38774.1 KI209939 ERL96114.1 NNAY01006739 OXU16364.1 KK117683 KFM71119.1 EZ424043 ADD20319.1 GBZX01002961 JAG89779.1 CCAG010016647 AFYH01050644 APGK01054147 APGK01054148 KB741247 ENN72041.1 MCFN01000098 OXB65288.1 GFDF01009793 JAV04291.1 KK739026 KFP38078.1 DQ216086 ACH45456.1 KL872353 KGL95213.1 KK589767 KFV99369.1 KL463494 KFV14699.1 KK612369 KFV46875.1 KL217789 KFO98727.1 KL410274 KFQ95189.1 KK735750 KFR15613.1 KL482444 KFO08445.1 KL328484 KFP57144.1 KK829451 KFP75327.1 KL227546 KFV03540.1 KK502098 KFP20631.1 KK451973 KFQ74098.1 AKCR02000089 PKK21165.1 KL225058 KFW67059.1 KK367895 KFV42015.1 KL349003 KFZ57640.1 KK569593 KFW06522.1 KL205870 KFV75576.1 KK659809 KFQ07434.1 KL226300 KFM11387.1 KB743085 EOB01507.1 DQ216084 DQ216085 ACH45454.1 KL435359 KFW92697.1 KK718435 KFO56327.1 AAPE02006101 KE161960 EPQ06097.1 AADN05000527 AGTP01046508 HAAF01000217 CCP72043.1
KPI98498.1 JTDY01004718 KOB67840.1 JTDY01005705 KOB66719.1 KZ288199 PBC33636.1 GFTR01000906 JAW15520.1 GBGD01003657 JAC85232.1 GBBI01000798 JAC17914.1 GECL01000260 JAP05864.1 PYGN01000332 PSN48374.1 KQ434803 KZC05909.1 KK852976 KDR13050.1 GDKW01002472 JAI54123.1 ACPB03020263 GAHY01002106 JAA75404.1 KQ971343 KYB27308.1 KQ767877 OAD53281.1 GL448858 EFN83647.1 GGFK01012352 MBW45673.1 KQ977234 KYN04703.1 KQ979039 KYN23334.1 GL888033 EGI69104.1 KQ981909 KYN33205.1 ADTU01000887 KQ983012 KYQ48371.1 APCN01002811 GGFM01005636 MBW26387.1 ADMH02000362 ETN66831.1 LBMM01004382 KMQ92481.1 GL436716 EFN71590.1 AAAB01008900 EAA09386.5 GGFJ01011289 MBW60430.1 KK107321 QOIP01000002 EZA52696.1 RLU25393.1 KQ414688 KOC63708.1 AXCN02001020 AXCM01001015 GADI01007021 JAA66787.1 ABJB010972031 DS884446 EEC15166.1 NEVH01016057 PNF26369.1 AAZX01003377 UFQS01002630 UFQT01002630 SSX14405.1 SSX33815.1 JXUM01149326 KQ570750 KXJ68307.1 CR858529 IAAA01022778 LAA07509.1 NEVH01005296 PNF38774.1 KI209939 ERL96114.1 NNAY01006739 OXU16364.1 KK117683 KFM71119.1 EZ424043 ADD20319.1 GBZX01002961 JAG89779.1 CCAG010016647 AFYH01050644 APGK01054147 APGK01054148 KB741247 ENN72041.1 MCFN01000098 OXB65288.1 GFDF01009793 JAV04291.1 KK739026 KFP38078.1 DQ216086 ACH45456.1 KL872353 KGL95213.1 KK589767 KFV99369.1 KL463494 KFV14699.1 KK612369 KFV46875.1 KL217789 KFO98727.1 KL410274 KFQ95189.1 KK735750 KFR15613.1 KL482444 KFO08445.1 KL328484 KFP57144.1 KK829451 KFP75327.1 KL227546 KFV03540.1 KK502098 KFP20631.1 KK451973 KFQ74098.1 AKCR02000089 PKK21165.1 KL225058 KFW67059.1 KK367895 KFV42015.1 KL349003 KFZ57640.1 KK569593 KFW06522.1 KL205870 KFV75576.1 KK659809 KFQ07434.1 KL226300 KFM11387.1 KB743085 EOB01507.1 DQ216084 DQ216085 ACH45454.1 KL435359 KFW92697.1 KK718435 KFO56327.1 AAPE02006101 KE161960 EPQ06097.1 AADN05000527 AGTP01046508 HAAF01000217 CCP72043.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000037510
UP000242457
UP000005203
+ More
UP000245037 UP000076502 UP000027135 UP000015103 UP000007266 UP000008237 UP000078542 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000076407 UP000075902 UP000075840 UP000000673 UP000036403 UP000000311 UP000007062 UP000053097 UP000279307 UP000053825 UP000095300 UP000075886 UP000075883 UP000075900 UP000001555 UP000235965 UP000075885 UP000075881 UP000002358 UP000095301 UP000069940 UP000249989 UP000001595 UP000075920 UP000030742 UP000215335 UP000054359 UP000092444 UP000008672 UP000019118 UP000198323 UP000053858 UP000054308 UP000053283 UP000053605 UP000053119 UP000053872 UP000054081 UP000053584 UP000053286 UP000189705 UP000052976 UP000001074 UP000079721 UP000000539 UP000005215 UP000007646
UP000245037 UP000076502 UP000027135 UP000015103 UP000007266 UP000008237 UP000078542 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000076407 UP000075902 UP000075840 UP000000673 UP000036403 UP000000311 UP000007062 UP000053097 UP000279307 UP000053825 UP000095300 UP000075886 UP000075883 UP000075900 UP000001555 UP000235965 UP000075885 UP000075881 UP000002358 UP000095301 UP000069940 UP000249989 UP000001595 UP000075920 UP000030742 UP000215335 UP000054359 UP000092444 UP000008672 UP000019118 UP000198323 UP000053858 UP000054308 UP000053283 UP000053605 UP000053119 UP000053872 UP000054081 UP000053584 UP000053286 UP000189705 UP000052976 UP000001074 UP000079721 UP000000539 UP000005215 UP000007646
PRIDE
Pfam
Interpro
IPR008401
Apc13p
+ More
IPR029060 PIN-like_dom_sf
IPR006984 Fcf1/Utp23
IPR035896 AN1-like_Znf
IPR000058 Znf_AN1
IPR027409 GroEL-like_apical_dom_sf
IPR000306 Znf_FYVE
IPR002498 PInositol-4-P-5-kinase_core
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR017455 Znf_FYVE-rel
IPR027484 PInositol-4-P-5-kinase_N
IPR002423 Cpn60/TCP-1
IPR029060 PIN-like_dom_sf
IPR006984 Fcf1/Utp23
IPR035896 AN1-like_Znf
IPR000058 Znf_AN1
IPR027409 GroEL-like_apical_dom_sf
IPR000306 Znf_FYVE
IPR002498 PInositol-4-P-5-kinase_core
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR017455 Znf_FYVE-rel
IPR027484 PInositol-4-P-5-kinase_N
IPR002423 Cpn60/TCP-1
Gene 3D
ProteinModelPortal
H9J9B3
S4PHD2
A0A212EGU5
A0A194PZN4
A0A0L7KXH9
A0A0L7KUM8
+ More
A0A2A3EQZ0 A0A088AF80 A0A224Y328 A0A069DNR7 A0A023FA82 A0A0V0GDT8 A0A2P8YVU8 A0A154P1T5 A0A067QVD4 A0A0P4VP53 R4G309 A0A139WH34 A0A310S9R7 E2BKZ3 A0A2M4AY03 A0A151IKA6 A0A195EEQ7 F4W9W8 A0A195EY46 A0A158NZI5 A0A151WKR5 A0A182XEV2 A0A182UK85 A0A182HNX8 A0A2M3ZCW0 W5JRS3 A0A0J7KQC2 E2A4Q6 Q7PQ30 A0A2M4C550 A0A026WAL0 A0A0L7QYQ3 A0A1I8PIM1 A0A182QYI4 A0A182MLP5 A0A182RKW0 A0A0K8R6K0 B7Q8J4 A0A2J7QCQ4 A0A182P8S8 A0A182K7W3 K7JCE8 A0A1I8N725 A0A336LCX2 A0A182G703 Q5RBV4 A0A2L2YHC5 A0A182WK22 A0A2J7RD63 U4UPW5 A0A232EDF1 A0A087U180 D3TRJ1 A0A0C9RZU1 A0A1B0FP98 H3AY45 N6TUI7 A0A226NCN8 A0A1L8DCU8 A0A091KF47 B5G2E5 A0A0A0ANS5 A0A093I9H5 A0A093CP24 A0A093ERQ3 A0A091HUA8 A0A091UW42 A0A091WL26 A0A087V753 A0A091LLC4 A0A091MLG3 A0A093DNA7 A0A091KA51 A0A091TG81 A0A2I0LUR1 A0A093R7P2 A0A093GNC1 A0A094KGE7 A0A093J3S4 A0A093H1N3 A0A091QD37 A0A087RD33 R0JW70 B5G2E3 A0A1U7RN24 A0A093R9D4 A0A091EI49 G1QBV4 A0A1S2ZMX8 S7PDV3 A0A1D5PVV7 I3N837 G3SST0 U6CR27
A0A2A3EQZ0 A0A088AF80 A0A224Y328 A0A069DNR7 A0A023FA82 A0A0V0GDT8 A0A2P8YVU8 A0A154P1T5 A0A067QVD4 A0A0P4VP53 R4G309 A0A139WH34 A0A310S9R7 E2BKZ3 A0A2M4AY03 A0A151IKA6 A0A195EEQ7 F4W9W8 A0A195EY46 A0A158NZI5 A0A151WKR5 A0A182XEV2 A0A182UK85 A0A182HNX8 A0A2M3ZCW0 W5JRS3 A0A0J7KQC2 E2A4Q6 Q7PQ30 A0A2M4C550 A0A026WAL0 A0A0L7QYQ3 A0A1I8PIM1 A0A182QYI4 A0A182MLP5 A0A182RKW0 A0A0K8R6K0 B7Q8J4 A0A2J7QCQ4 A0A182P8S8 A0A182K7W3 K7JCE8 A0A1I8N725 A0A336LCX2 A0A182G703 Q5RBV4 A0A2L2YHC5 A0A182WK22 A0A2J7RD63 U4UPW5 A0A232EDF1 A0A087U180 D3TRJ1 A0A0C9RZU1 A0A1B0FP98 H3AY45 N6TUI7 A0A226NCN8 A0A1L8DCU8 A0A091KF47 B5G2E5 A0A0A0ANS5 A0A093I9H5 A0A093CP24 A0A093ERQ3 A0A091HUA8 A0A091UW42 A0A091WL26 A0A087V753 A0A091LLC4 A0A091MLG3 A0A093DNA7 A0A091KA51 A0A091TG81 A0A2I0LUR1 A0A093R7P2 A0A093GNC1 A0A094KGE7 A0A093J3S4 A0A093H1N3 A0A091QD37 A0A087RD33 R0JW70 B5G2E3 A0A1U7RN24 A0A093R9D4 A0A091EI49 G1QBV4 A0A1S2ZMX8 S7PDV3 A0A1D5PVV7 I3N837 G3SST0 U6CR27
PDB
5LCW
E-value=1.76859e-09,
Score=143
Ontologies
KEGG
GO
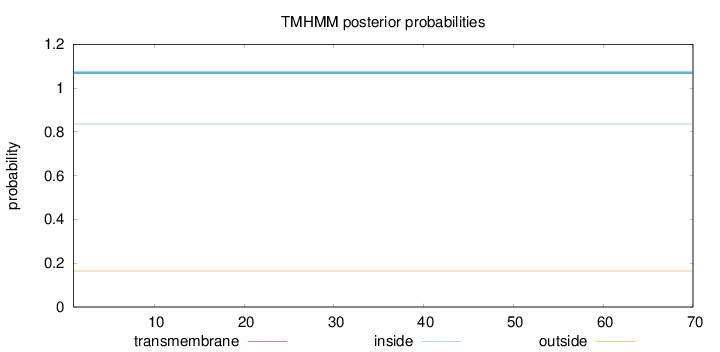
Topology
Subcellular location
Nucleus
Length:
70
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.83602
inside
1 - 70
Population Genetic Test Statistics
Pi
260.476347
Theta
202.812636
Tajima's D
0.903684
CLR
0.002506
CSRT
0.624918754062297
Interpretation
Uncertain
