Pre Gene Modal
BGIBMGA006105
Annotation
PREDICTED:_rRNA-processing_protein_UTP23_homolog_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.844 Nuclear Reliability : 2.444
Sequence
CDS
ATGAAAATAACACGTTACAAAAAAGTACAGAAATATCTAAAGTTTTATTACAACAATTATGGATTTCATCAGCCGTACCAAGTCCTTATCGATGGTACATTTTGTTTCGCTGCATTCAAAGAGCATATTAACATCAGAGAGCAAATTCCCAAGTACCTAAACAGCCAGGCTAAACTACTGACAACGAGATGTATAATTATAGAAACAGAAAAATTAGCAAAGAAAACACATGGAGCTCTCACAATATTAAAACAGTTTGGTATTCATGAATGTGGGCACAAAGAACCGGTGGCTGGATCTAAGTGCATAATGTCCATGATTGGTAAGAAGAATGAAAAGCATTATATCTTAGCTACCCAGGATCGAGATCTGCAATCCAAATTAAGGGAAAAACCTGGAGTTCCTCTGCTATATCTACATAACAAGTCACCAACATTAGAGAAACCAAGTAGTGTGAGCTTTGACAAAGCAGGTCAAGCTCTGGAAACCAATGAACATATTTTTATCACTGAGAATCAGAGTGAAACTTTGAAGCAGATGAAAAAGAAATTAGGTGTTGAAGAAAAGGTTGAAGAACTGAATGTCCCTATTAGGAAGAAAAAGCCCCATAACCCTAATCCTTTGTCTTGTAAGAAAAAGAAAAATAAACCAAGCAGACAAGGAACAAAAAAAGAAAATGTGACTGAAGGAAAAGTCCGGAAAAGGAAGAAAAATAAGTTTCCGAAGCATTTAAAAAATGAACTTAAAAGTGTTATTAGTCAGTAA
Protein
MKITRYKKVQKYLKFYYNNYGFHQPYQVLIDGTFCFAAFKEHINIREQIPKYLNSQAKLLTTRCIIIETEKLAKKTHGALTILKQFGIHECGHKEPVAGSKCIMSMIGKKNEKHYILATQDRDLQSKLREKPGVPLLYLHNKSPTLEKPSSVSFDKAGQALETNEHIFITENQSETLKQMKKKLGVEEKVEELNVPIRKKKPHNPNPLSCKKKKNKPSRQGTKKENVTEGKVRKRKKNKFPKHLKNELKSVISQ
Summary
Uniprot
A0A2A4IYG6
A0A194RBH8
A0A2H1W6A3
A0A1E1WV83
H9J9B3
A0A0L7KXH9
+ More
A0A0L7KUM8 A0A2W1BCT5 A0A1B6DG47 A0A212EGU5 A0A0T6AY60 A0A232EKB1 K7INF5 A0A0P4VHJ4 R4G7R4 A0A1W4WQC1 A0A0A9WPS1 A0A0K8T081 A0A0V0GCF8 N6T502 J3JTY8 A0A067RPN8 A0A131XBY7 F4WSE9 E9HKH2 T1IRT6 A0A151X373 A0A195ESU1 A0A2P8Y797 E2AXF9 A0A195D409 A0A026W1V6 A0A195EET9 A0A1E1X4K9 A0A023GJM5 G3MLL2 A0A0P5G9S5 A0A1E1XRG1 L7M7Q7 A0A131YNQ0 A0A1B6MHJ1 A0A023FXQ9 A0A1Y1N120 A0A224Z968 E9IYH9 A0A0P5P328 A0A0P5BKM4 A0A195BQ65 A0A158NTM1 A0A131YA61 A0A0P4Y3Q4 A0A224ZB73 A0A0P5KGW7 E2BKU6 A0A0P6IJA3 B7Q827 A0A0P5XEP8 A0A0P5XUW2 K7G317 A0A0K8TM71 A0A132A1R7 A0A087ZMT9 V5H970 A0A154PGA8 A0A2R5LI14 A0A2L2YCA2 A0A336LGU9 A0A293LUE8 A0A1Z5L6I3 A0A1D1VYT5 A0A182Q744 A0A0L0C472 A0A2D0PWB9 A0A3M0L6W9 B0WE53 A0A226N5N0 U3JHY9 U5EVD5 W4YZ89 A0A1U7U5W5 A0A2I2UIZ8 G1NIM9 A0A1D5PZG8 A0A1I8N8A7 A0A182UCR8 A0A1A9UMG5 B3MKE6 A0A182ICC4 A0A182XLA0 B4LVK0
A0A0L7KUM8 A0A2W1BCT5 A0A1B6DG47 A0A212EGU5 A0A0T6AY60 A0A232EKB1 K7INF5 A0A0P4VHJ4 R4G7R4 A0A1W4WQC1 A0A0A9WPS1 A0A0K8T081 A0A0V0GCF8 N6T502 J3JTY8 A0A067RPN8 A0A131XBY7 F4WSE9 E9HKH2 T1IRT6 A0A151X373 A0A195ESU1 A0A2P8Y797 E2AXF9 A0A195D409 A0A026W1V6 A0A195EET9 A0A1E1X4K9 A0A023GJM5 G3MLL2 A0A0P5G9S5 A0A1E1XRG1 L7M7Q7 A0A131YNQ0 A0A1B6MHJ1 A0A023FXQ9 A0A1Y1N120 A0A224Z968 E9IYH9 A0A0P5P328 A0A0P5BKM4 A0A195BQ65 A0A158NTM1 A0A131YA61 A0A0P4Y3Q4 A0A224ZB73 A0A0P5KGW7 E2BKU6 A0A0P6IJA3 B7Q827 A0A0P5XEP8 A0A0P5XUW2 K7G317 A0A0K8TM71 A0A132A1R7 A0A087ZMT9 V5H970 A0A154PGA8 A0A2R5LI14 A0A2L2YCA2 A0A336LGU9 A0A293LUE8 A0A1Z5L6I3 A0A1D1VYT5 A0A182Q744 A0A0L0C472 A0A2D0PWB9 A0A3M0L6W9 B0WE53 A0A226N5N0 U3JHY9 U5EVD5 W4YZ89 A0A1U7U5W5 A0A2I2UIZ8 G1NIM9 A0A1D5PZG8 A0A1I8N8A7 A0A182UCR8 A0A1A9UMG5 B3MKE6 A0A182ICC4 A0A182XLA0 B4LVK0
Pubmed
26354079
19121390
26227816
28756777
22118469
28648823
+ More
20075255 27129103 25401762 23537049 22516182 24845553 28049606 21719571 21292972 29403074 20798317 24508170 28503490 22216098 29209593 25576852 26830274 28004739 28797301 21282665 21347285 17381049 26369729 26555130 25765539 26561354 28528879 27649274 26108605 17975172 20838655 15592404 25315136 17994087
20075255 27129103 25401762 23537049 22516182 24845553 28049606 21719571 21292972 29403074 20798317 24508170 28503490 22216098 29209593 25576852 26830274 28004739 28797301 21282665 21347285 17381049 26369729 26555130 25765539 26561354 28528879 27649274 26108605 17975172 20838655 15592404 25315136 17994087
EMBL
NWSH01004520
PCG64997.1
KQ460644
KPJ13256.1
ODYU01006603
SOQ48588.1
+ More
GDQN01000187 JAT90867.1 BABH01004417 JTDY01004718 KOB67840.1 JTDY01005705 KOB66719.1 KZ150155 PZC72788.1 GEDC01012651 JAS24647.1 AGBW02015019 OWR40704.1 LJIG01022548 KRT79994.1 NNAY01003819 OXU18804.1 GDKW01002632 JAI53963.1 GAHY01002267 JAA75243.1 GBHO01033152 JAG10452.1 GBRD01007251 JAG58570.1 GECL01001074 JAP05050.1 APGK01052432 KB741212 KB632279 ENN72723.1 ERL91277.1 BT126698 AEE61660.1 KK852498 KDR22590.1 GEFH01004951 JAP63630.1 GL888321 EGI62866.1 GL732669 EFX67759.1 JH431385 KQ982562 KYQ54851.1 KQ981993 KYN30982.1 PYGN01000842 PSN40120.1 GL443549 EFN61883.1 KQ976881 KYN07622.1 KK107549 EZA49039.1 KQ978983 KYN26753.1 GFAC01004978 JAT94210.1 GBBM01002175 JAC33243.1 JO842763 AEO34380.1 GDIQ01245457 JAK06268.1 GFAA01001913 JAU01522.1 GACK01005846 JAA59188.1 GEDV01007663 JAP80894.1 GEBQ01004584 JAT35393.1 GBBL01001850 JAC25470.1 GEZM01016841 GEZM01016839 GEZM01016838 GEZM01016837 JAV91128.1 GFPF01013005 MAA24151.1 GL767018 EFZ14337.1 GDIQ01134493 JAL17233.1 GDIP01198204 JAJ25198.1 KQ976424 KYM88667.1 ADTU01026032 GEFM01000368 JAP75428.1 GDIP01233990 JAI89411.1 GFPF01013004 MAA24150.1 GDIQ01189665 JAK62060.1 GL448826 EFN83695.1 GDIQ01020982 JAN73755.1 ABJB010318398 DS880154 EEC14999.1 GDIP01074762 JAM28953.1 GDIP01067168 JAM36547.1 AGCU01137658 GDAI01002149 JAI15454.1 JXLN01009946 KPM04759.1 GANP01010754 JAB73714.1 KQ434899 KZC10883.1 GGLE01004929 MBY09055.1 IAAA01009983 LAA04805.1 UFQS01000004 UFQT01000004 SSW96815.1 SSX17202.1 GFWV01006678 MAA31408.1 GFJQ02004011 JAW02959.1 BDGG01000013 GAV06570.1 AXCN02000805 JRES01000934 KNC27085.1 QRBI01000104 RMC14977.1 DS231904 EDS45229.1 MCFN01000193 OXB62905.1 AGTO01020541 GANO01001926 JAB57945.1 AAGJ04151527 AANG04003518 AADN05000663 CH902620 EDV32530.1 APCN01000793 CH940649 EDW64394.1
GDQN01000187 JAT90867.1 BABH01004417 JTDY01004718 KOB67840.1 JTDY01005705 KOB66719.1 KZ150155 PZC72788.1 GEDC01012651 JAS24647.1 AGBW02015019 OWR40704.1 LJIG01022548 KRT79994.1 NNAY01003819 OXU18804.1 GDKW01002632 JAI53963.1 GAHY01002267 JAA75243.1 GBHO01033152 JAG10452.1 GBRD01007251 JAG58570.1 GECL01001074 JAP05050.1 APGK01052432 KB741212 KB632279 ENN72723.1 ERL91277.1 BT126698 AEE61660.1 KK852498 KDR22590.1 GEFH01004951 JAP63630.1 GL888321 EGI62866.1 GL732669 EFX67759.1 JH431385 KQ982562 KYQ54851.1 KQ981993 KYN30982.1 PYGN01000842 PSN40120.1 GL443549 EFN61883.1 KQ976881 KYN07622.1 KK107549 EZA49039.1 KQ978983 KYN26753.1 GFAC01004978 JAT94210.1 GBBM01002175 JAC33243.1 JO842763 AEO34380.1 GDIQ01245457 JAK06268.1 GFAA01001913 JAU01522.1 GACK01005846 JAA59188.1 GEDV01007663 JAP80894.1 GEBQ01004584 JAT35393.1 GBBL01001850 JAC25470.1 GEZM01016841 GEZM01016839 GEZM01016838 GEZM01016837 JAV91128.1 GFPF01013005 MAA24151.1 GL767018 EFZ14337.1 GDIQ01134493 JAL17233.1 GDIP01198204 JAJ25198.1 KQ976424 KYM88667.1 ADTU01026032 GEFM01000368 JAP75428.1 GDIP01233990 JAI89411.1 GFPF01013004 MAA24150.1 GDIQ01189665 JAK62060.1 GL448826 EFN83695.1 GDIQ01020982 JAN73755.1 ABJB010318398 DS880154 EEC14999.1 GDIP01074762 JAM28953.1 GDIP01067168 JAM36547.1 AGCU01137658 GDAI01002149 JAI15454.1 JXLN01009946 KPM04759.1 GANP01010754 JAB73714.1 KQ434899 KZC10883.1 GGLE01004929 MBY09055.1 IAAA01009983 LAA04805.1 UFQS01000004 UFQT01000004 SSW96815.1 SSX17202.1 GFWV01006678 MAA31408.1 GFJQ02004011 JAW02959.1 BDGG01000013 GAV06570.1 AXCN02000805 JRES01000934 KNC27085.1 QRBI01000104 RMC14977.1 DS231904 EDS45229.1 MCFN01000193 OXB62905.1 AGTO01020541 GANO01001926 JAB57945.1 AAGJ04151527 AANG04003518 AADN05000663 CH902620 EDV32530.1 APCN01000793 CH940649 EDW64394.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000037510
UP000007151
UP000215335
+ More
UP000002358 UP000192223 UP000019118 UP000030742 UP000027135 UP000007755 UP000000305 UP000075809 UP000078541 UP000245037 UP000000311 UP000078542 UP000053097 UP000078492 UP000078540 UP000005205 UP000008237 UP000001555 UP000007267 UP000005203 UP000076502 UP000186922 UP000075886 UP000037069 UP000221080 UP000269221 UP000002320 UP000198323 UP000016665 UP000007110 UP000189704 UP000011712 UP000001645 UP000000539 UP000095301 UP000075902 UP000078200 UP000007801 UP000075840 UP000076407 UP000008792
UP000002358 UP000192223 UP000019118 UP000030742 UP000027135 UP000007755 UP000000305 UP000075809 UP000078541 UP000245037 UP000000311 UP000078542 UP000053097 UP000078492 UP000078540 UP000005205 UP000008237 UP000001555 UP000007267 UP000005203 UP000076502 UP000186922 UP000075886 UP000037069 UP000221080 UP000269221 UP000002320 UP000198323 UP000016665 UP000007110 UP000189704 UP000011712 UP000001645 UP000000539 UP000095301 UP000075902 UP000078200 UP000007801 UP000075840 UP000076407 UP000008792
PRIDE
SUPFAM
SSF88723
SSF88723
ProteinModelPortal
A0A2A4IYG6
A0A194RBH8
A0A2H1W6A3
A0A1E1WV83
H9J9B3
A0A0L7KXH9
+ More
A0A0L7KUM8 A0A2W1BCT5 A0A1B6DG47 A0A212EGU5 A0A0T6AY60 A0A232EKB1 K7INF5 A0A0P4VHJ4 R4G7R4 A0A1W4WQC1 A0A0A9WPS1 A0A0K8T081 A0A0V0GCF8 N6T502 J3JTY8 A0A067RPN8 A0A131XBY7 F4WSE9 E9HKH2 T1IRT6 A0A151X373 A0A195ESU1 A0A2P8Y797 E2AXF9 A0A195D409 A0A026W1V6 A0A195EET9 A0A1E1X4K9 A0A023GJM5 G3MLL2 A0A0P5G9S5 A0A1E1XRG1 L7M7Q7 A0A131YNQ0 A0A1B6MHJ1 A0A023FXQ9 A0A1Y1N120 A0A224Z968 E9IYH9 A0A0P5P328 A0A0P5BKM4 A0A195BQ65 A0A158NTM1 A0A131YA61 A0A0P4Y3Q4 A0A224ZB73 A0A0P5KGW7 E2BKU6 A0A0P6IJA3 B7Q827 A0A0P5XEP8 A0A0P5XUW2 K7G317 A0A0K8TM71 A0A132A1R7 A0A087ZMT9 V5H970 A0A154PGA8 A0A2R5LI14 A0A2L2YCA2 A0A336LGU9 A0A293LUE8 A0A1Z5L6I3 A0A1D1VYT5 A0A182Q744 A0A0L0C472 A0A2D0PWB9 A0A3M0L6W9 B0WE53 A0A226N5N0 U3JHY9 U5EVD5 W4YZ89 A0A1U7U5W5 A0A2I2UIZ8 G1NIM9 A0A1D5PZG8 A0A1I8N8A7 A0A182UCR8 A0A1A9UMG5 B3MKE6 A0A182ICC4 A0A182XLA0 B4LVK0
A0A0L7KUM8 A0A2W1BCT5 A0A1B6DG47 A0A212EGU5 A0A0T6AY60 A0A232EKB1 K7INF5 A0A0P4VHJ4 R4G7R4 A0A1W4WQC1 A0A0A9WPS1 A0A0K8T081 A0A0V0GCF8 N6T502 J3JTY8 A0A067RPN8 A0A131XBY7 F4WSE9 E9HKH2 T1IRT6 A0A151X373 A0A195ESU1 A0A2P8Y797 E2AXF9 A0A195D409 A0A026W1V6 A0A195EET9 A0A1E1X4K9 A0A023GJM5 G3MLL2 A0A0P5G9S5 A0A1E1XRG1 L7M7Q7 A0A131YNQ0 A0A1B6MHJ1 A0A023FXQ9 A0A1Y1N120 A0A224Z968 E9IYH9 A0A0P5P328 A0A0P5BKM4 A0A195BQ65 A0A158NTM1 A0A131YA61 A0A0P4Y3Q4 A0A224ZB73 A0A0P5KGW7 E2BKU6 A0A0P6IJA3 B7Q827 A0A0P5XEP8 A0A0P5XUW2 K7G317 A0A0K8TM71 A0A132A1R7 A0A087ZMT9 V5H970 A0A154PGA8 A0A2R5LI14 A0A2L2YCA2 A0A336LGU9 A0A293LUE8 A0A1Z5L6I3 A0A1D1VYT5 A0A182Q744 A0A0L0C472 A0A2D0PWB9 A0A3M0L6W9 B0WE53 A0A226N5N0 U3JHY9 U5EVD5 W4YZ89 A0A1U7U5W5 A0A2I2UIZ8 G1NIM9 A0A1D5PZG8 A0A1I8N8A7 A0A182UCR8 A0A1A9UMG5 B3MKE6 A0A182ICC4 A0A182XLA0 B4LVK0
PDB
4MJ7
E-value=4.62202e-16,
Score=204
Ontologies
GO
Topology
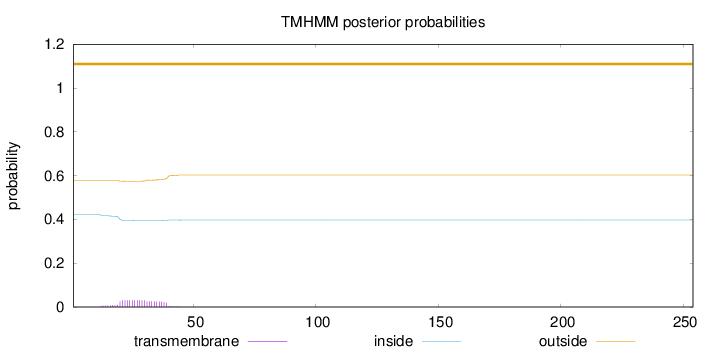
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.62084
Exp number, first 60 AAs:
0.62078
Total prob of N-in:
0.42314
outside
1 - 254
Population Genetic Test Statistics
Pi
258.110099
Theta
193.874929
Tajima's D
0.867581
CLR
0.002506
CSRT
0.625068746562672
Interpretation
Uncertain
