Gene
KWMTBOMO01662 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006045
Annotation
small_GTP_binding_protein_RAB5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.416
Sequence
CDS
ATGGCGACAAACAGGACCGGAGCCGCACAGAGGCCCAATGGCGCTCCACAGACAAAAGTCTGCCAGTTCAAGCTGGTGTTGTTGGGCGAGTCTGCGGTAGGCAAGTCCTCGCTGGTGCTCCGATTCGTCAAGGGACAGTTCCACGAGTACCAGGAGAGCACCATCGGGGCTGCGTTCCTCACCCAGGCCATTCGATTCGACGACACCACCGTTAAATTCGAGATCTGGGATACAGCTGGACAGGAGAGGTATCACAGCTTAGCGCCGATGTACTACAGAGGAGCCCAATCTGCTATTGTAGTATATGATATAACTAATCAGGACACATTCGGTCGCGCCAAGAACTGGGTGAAGGAACTCCAACGGCAGGCCTCTCCGTCGATAGTGATCGCTCTGGCGGGCAACAAGAGCGACCTCGCAGCCAAGCGCATGGTCGAGTTCGAGGAGGCGCAGGCCTACGCAGACGAGAACGGGCTGCTCTTCATGGAGACCAGCGCCAAGACGGCCATGAACGTCAACGATATATTCCTCGCTATAGCGAACAAGTTACCAAAGAGCGAGGCGGGGGCGGCGGGCGCGGCGGGGGGCGGGCGGCGCCTGGCGGACGGCGAGCAGCCGCGACCCTCGCAGTGCTGCAAGTGA
Protein
MATNRTGAAQRPNGAPQTKVCQFKLVLLGESAVGKSSLVLRFVKGQFHEYQESTIGAAFLTQAIRFDDTTVKFEIWDTAGQERYHSLAPMYYRGAQSAIVVYDITNQDTFGRAKNWVKELQRQASPSIVIALAGNKSDLAAKRMVEFEEAQAYADENGLLFMETSAKTAMNVNDIFLAIANKLPKSEAGAAGAAGGGRRLADGEQPRPSQCCK
Summary
Uniprot
Q19N36
A0A194PYU3
A0A2H1W672
A0A194R6G7
A0A212EGT7
S4PZU7
+ More
D6WJQ8 A0A1W4XB07 N6U3V1 A0A224XXU2 A0A0P4VM87 A0A0V0G6J0 A0A023F9Y3 A0A069DQS6 R4FPV9 A0A023GG88 A0A224Z7I0 A0A023FJI3 L7M4V0 A0A023FV39 A0A131XDE5 A0A1E1X231 A0A1D2N312 E2BP20 A0A1Y1MGW0 V5GWI1 A0A023GF17 A0A2R5L9F5 A0A195E703 F4X6V7 A0A151XC02 A0A2T7PWI0 E9J660 E1ZYZ0 A0A026W7Q2 A0A0J7KXD6 A0A158NFX1 A0A195B2M4 T1J5J1 V5HZ27 A0A2J7Q0P2 A0A067RWS2 V9IG87 K1R1Q8 V3ZPS1 A0A1Z5KWW0 A0A310SMQ2 A0A154PPM4 A0A0L7QJX7 A0A2I3TG75 A0A210QN54 A0A087UC73 F7HL78 A0A293M4T7 A0A2K6APR7 A0A2K5UNN5 A0A2K5YYA3 A0A2I3MUN9 A0A232FDK6 K7J3J9 A0A2I9LPI3 A0A226F484 I3MFP2 A0A1B6GSF6 R7U9J3 A0A0L8HMP5 A0A1X7UJA5 A7RG79 A0A0K8TPC0 A0A1U7TQF4 A0A1B6MKB1 A0A1B6IDA1 A0A2K6APS0 A0A2K6JUQ8 A0A2K5YY97 A0A0D9QZ02 G7PIG1 A0A2K6QHW7 G7N7A8 A0A384DTH5 D2H337 A0A1A6GXF5 A0A250XWG6 A0A286X8R6 A0A091D0Q5 A0A1S3FN70 B2RPS1 A0A1U7QS77 E2QW69 G1S735 A0A341ATE0 A0A2Y9R4C6 A0A2U3ZYN7 A0A2R9B4V1 A0A2Y9FUJ5 A0A2Y9GU78 A0A2U3XBJ7 A0A2Y9L023 A0A2U3W147 L8Y6K1
D6WJQ8 A0A1W4XB07 N6U3V1 A0A224XXU2 A0A0P4VM87 A0A0V0G6J0 A0A023F9Y3 A0A069DQS6 R4FPV9 A0A023GG88 A0A224Z7I0 A0A023FJI3 L7M4V0 A0A023FV39 A0A131XDE5 A0A1E1X231 A0A1D2N312 E2BP20 A0A1Y1MGW0 V5GWI1 A0A023GF17 A0A2R5L9F5 A0A195E703 F4X6V7 A0A151XC02 A0A2T7PWI0 E9J660 E1ZYZ0 A0A026W7Q2 A0A0J7KXD6 A0A158NFX1 A0A195B2M4 T1J5J1 V5HZ27 A0A2J7Q0P2 A0A067RWS2 V9IG87 K1R1Q8 V3ZPS1 A0A1Z5KWW0 A0A310SMQ2 A0A154PPM4 A0A0L7QJX7 A0A2I3TG75 A0A210QN54 A0A087UC73 F7HL78 A0A293M4T7 A0A2K6APR7 A0A2K5UNN5 A0A2K5YYA3 A0A2I3MUN9 A0A232FDK6 K7J3J9 A0A2I9LPI3 A0A226F484 I3MFP2 A0A1B6GSF6 R7U9J3 A0A0L8HMP5 A0A1X7UJA5 A7RG79 A0A0K8TPC0 A0A1U7TQF4 A0A1B6MKB1 A0A1B6IDA1 A0A2K6APS0 A0A2K6JUQ8 A0A2K5YY97 A0A0D9QZ02 G7PIG1 A0A2K6QHW7 G7N7A8 A0A384DTH5 D2H337 A0A1A6GXF5 A0A250XWG6 A0A286X8R6 A0A091D0Q5 A0A1S3FN70 B2RPS1 A0A1U7QS77 E2QW69 G1S735 A0A341ATE0 A0A2Y9R4C6 A0A2U3ZYN7 A0A2R9B4V1 A0A2Y9FUJ5 A0A2Y9GU78 A0A2U3XBJ7 A0A2Y9L023 A0A2U3W147 L8Y6K1
Pubmed
19121390
26354079
22118469
23622113
18362917
19820115
+ More
23537049 27129103 25474469 26334808 28797301 25576852 28049606 28503490 27289101 20798317 28004739 21719571 21282665 24508170 30249741 21347285 25765539 24845553 22992520 23254933 28528879 16136131 28812685 17431167 28648823 20075255 29248469 30926861 17615350 26369729 22002653 25362486 25319552 24813606 20010809 28087693 21993624 12040188 15489334 21183079 16341006 22722832 23385571
23537049 27129103 25474469 26334808 28797301 25576852 28049606 28503490 27289101 20798317 28004739 21719571 21282665 24508170 30249741 21347285 25765539 24845553 22992520 23254933 28528879 16136131 28812685 17431167 28648823 20075255 29248469 30926861 17615350 26369729 22002653 25362486 25319552 24813606 20010809 28087693 21993624 12040188 15489334 21183079 16341006 22722832 23385571
EMBL
BABH01004414
BABH01004415
BABH01004416
DQ517934
ABF66621.1
KQ459584
+ More
KPI98496.1 ODYU01006603 SOQ48589.1 KQ460644 KPJ13257.1 AGBW02015019 OWR40705.1 GAIX01000918 JAA91642.1 KQ971343 EFA04701.1 APGK01040754 KB740984 KB631904 ENN76270.1 ERL87098.1 GFTR01003585 JAW12841.1 GDKW01000962 JAI55633.1 GECL01003239 JAP02885.1 GBBI01000502 JAC18210.1 GBGD01002897 JAC85992.1 ACPB03001109 GAHY01000694 JAA76816.1 GBBM01002481 JAC32937.1 GFPF01011234 MAA22380.1 GBBK01003639 JAC20843.1 GACK01006137 JAA58897.1 GBBL01002780 JAC24540.1 GEFH01003984 JAP64597.1 GFAC01005865 JAT93323.1 LJIJ01000288 ODM99305.1 GL449511 EFN82612.1 GEZM01035708 GEZM01035707 JAV83116.1 GALX01003868 JAB64598.1 GBBM01002732 JAC32686.1 GGLE01001979 MBY06105.1 KQ979568 KYN20876.1 GL888818 EGI57889.1 KQ982314 KYQ57916.1 PZQS01000001 PVD37747.1 GL768221 EFZ11732.1 GL435242 EFN73680.1 KK107373 QOIP01000004 EZA51651.1 RLU23292.1 LBMM01002177 KMQ95177.1 ADTU01014821 KQ976662 KYM78542.1 JH431865 GANP01000324 JAB84144.1 NEVH01019965 PNF22129.1 KK852415 KDR24359.1 JR045964 AEY60098.1 JH816269 EKC27651.1 KB203470 ESO84515.1 GFJQ02007391 JAV99578.1 KQ763450 OAD55058.1 KQ434981 KZC13120.1 KQ415041 KOC58894.1 AACZ04012961 NEDP02002754 OWF50173.1 KK119171 KFM74962.1 JSUE03006924 GFWV01009256 MAA33985.1 AQIA01011136 AQIA01011137 AQIA01011138 AQIA01011139 AQIA01011140 AHZZ02002258 NNAY01000390 OXU28722.1 GFWZ01000305 MBW20295.1 LNIX01000001 MH637938 OXA64589.1 QBH73328.1 AGTP01085300 AGTP01085301 AGTP01085302 AGTP01085303 GECZ01004409 JAS65360.1 AMQN01008683 KB303737 ELU02811.1 KQ417851 KOF90050.1 DS469509 EDO49482.1 GDAI01001612 JAI15991.1 GEBQ01003570 JAT36407.1 GECU01022788 GECU01018643 JAS84918.1 JAS89063.1 AQIB01023024 AQIB01023025 CM001286 EHH66384.1 JU321692 JU471906 JV043549 CM001263 AFE65448.1 AFH28710.1 AFI33620.1 EHH20843.1 GL192457 EFB28835.1 LZPO01066261 OBS71018.1 GFFV01004274 JAV35671.1 AAKN02014842 KN123543 KFO24582.1 BC137574 BC137575 CH466578 AAI37575.1 AAI37576.1 EDL24597.1 AAEX03006917 ADFV01002493 ADFV01002494 AJFE02087842 AJFE02087843 AJFE02087844 AJFE02087845 KB364768 ELV12033.1
KPI98496.1 ODYU01006603 SOQ48589.1 KQ460644 KPJ13257.1 AGBW02015019 OWR40705.1 GAIX01000918 JAA91642.1 KQ971343 EFA04701.1 APGK01040754 KB740984 KB631904 ENN76270.1 ERL87098.1 GFTR01003585 JAW12841.1 GDKW01000962 JAI55633.1 GECL01003239 JAP02885.1 GBBI01000502 JAC18210.1 GBGD01002897 JAC85992.1 ACPB03001109 GAHY01000694 JAA76816.1 GBBM01002481 JAC32937.1 GFPF01011234 MAA22380.1 GBBK01003639 JAC20843.1 GACK01006137 JAA58897.1 GBBL01002780 JAC24540.1 GEFH01003984 JAP64597.1 GFAC01005865 JAT93323.1 LJIJ01000288 ODM99305.1 GL449511 EFN82612.1 GEZM01035708 GEZM01035707 JAV83116.1 GALX01003868 JAB64598.1 GBBM01002732 JAC32686.1 GGLE01001979 MBY06105.1 KQ979568 KYN20876.1 GL888818 EGI57889.1 KQ982314 KYQ57916.1 PZQS01000001 PVD37747.1 GL768221 EFZ11732.1 GL435242 EFN73680.1 KK107373 QOIP01000004 EZA51651.1 RLU23292.1 LBMM01002177 KMQ95177.1 ADTU01014821 KQ976662 KYM78542.1 JH431865 GANP01000324 JAB84144.1 NEVH01019965 PNF22129.1 KK852415 KDR24359.1 JR045964 AEY60098.1 JH816269 EKC27651.1 KB203470 ESO84515.1 GFJQ02007391 JAV99578.1 KQ763450 OAD55058.1 KQ434981 KZC13120.1 KQ415041 KOC58894.1 AACZ04012961 NEDP02002754 OWF50173.1 KK119171 KFM74962.1 JSUE03006924 GFWV01009256 MAA33985.1 AQIA01011136 AQIA01011137 AQIA01011138 AQIA01011139 AQIA01011140 AHZZ02002258 NNAY01000390 OXU28722.1 GFWZ01000305 MBW20295.1 LNIX01000001 MH637938 OXA64589.1 QBH73328.1 AGTP01085300 AGTP01085301 AGTP01085302 AGTP01085303 GECZ01004409 JAS65360.1 AMQN01008683 KB303737 ELU02811.1 KQ417851 KOF90050.1 DS469509 EDO49482.1 GDAI01001612 JAI15991.1 GEBQ01003570 JAT36407.1 GECU01022788 GECU01018643 JAS84918.1 JAS89063.1 AQIB01023024 AQIB01023025 CM001286 EHH66384.1 JU321692 JU471906 JV043549 CM001263 AFE65448.1 AFH28710.1 AFI33620.1 EHH20843.1 GL192457 EFB28835.1 LZPO01066261 OBS71018.1 GFFV01004274 JAV35671.1 AAKN02014842 KN123543 KFO24582.1 BC137574 BC137575 CH466578 AAI37575.1 AAI37576.1 EDL24597.1 AAEX03006917 ADFV01002493 ADFV01002494 AJFE02087842 AJFE02087843 AJFE02087844 AJFE02087845 KB364768 ELV12033.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000007266
UP000192223
+ More
UP000019118 UP000030742 UP000015103 UP000094527 UP000008237 UP000078492 UP000007755 UP000075809 UP000245119 UP000000311 UP000053097 UP000279307 UP000036403 UP000005205 UP000078540 UP000235965 UP000027135 UP000005408 UP000030746 UP000076502 UP000053825 UP000002277 UP000242188 UP000054359 UP000006718 UP000233120 UP000233100 UP000233140 UP000028761 UP000215335 UP000002358 UP000198287 UP000005215 UP000014760 UP000053454 UP000007879 UP000001593 UP000189704 UP000233180 UP000029965 UP000009130 UP000233200 UP000261680 UP000291021 UP000092124 UP000005447 UP000028990 UP000081671 UP000189706 UP000002254 UP000001073 UP000252040 UP000248480 UP000245320 UP000240080 UP000248484 UP000248481 UP000245341 UP000248482 UP000245340 UP000011518
UP000019118 UP000030742 UP000015103 UP000094527 UP000008237 UP000078492 UP000007755 UP000075809 UP000245119 UP000000311 UP000053097 UP000279307 UP000036403 UP000005205 UP000078540 UP000235965 UP000027135 UP000005408 UP000030746 UP000076502 UP000053825 UP000002277 UP000242188 UP000054359 UP000006718 UP000233120 UP000233100 UP000233140 UP000028761 UP000215335 UP000002358 UP000198287 UP000005215 UP000014760 UP000053454 UP000007879 UP000001593 UP000189704 UP000233180 UP000029965 UP000009130 UP000233200 UP000261680 UP000291021 UP000092124 UP000005447 UP000028990 UP000081671 UP000189706 UP000002254 UP000001073 UP000252040 UP000248480 UP000245320 UP000240080 UP000248484 UP000248481 UP000245341 UP000248482 UP000245340 UP000011518
Pfam
PF00071 Ras
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q19N36
A0A194PYU3
A0A2H1W672
A0A194R6G7
A0A212EGT7
S4PZU7
+ More
D6WJQ8 A0A1W4XB07 N6U3V1 A0A224XXU2 A0A0P4VM87 A0A0V0G6J0 A0A023F9Y3 A0A069DQS6 R4FPV9 A0A023GG88 A0A224Z7I0 A0A023FJI3 L7M4V0 A0A023FV39 A0A131XDE5 A0A1E1X231 A0A1D2N312 E2BP20 A0A1Y1MGW0 V5GWI1 A0A023GF17 A0A2R5L9F5 A0A195E703 F4X6V7 A0A151XC02 A0A2T7PWI0 E9J660 E1ZYZ0 A0A026W7Q2 A0A0J7KXD6 A0A158NFX1 A0A195B2M4 T1J5J1 V5HZ27 A0A2J7Q0P2 A0A067RWS2 V9IG87 K1R1Q8 V3ZPS1 A0A1Z5KWW0 A0A310SMQ2 A0A154PPM4 A0A0L7QJX7 A0A2I3TG75 A0A210QN54 A0A087UC73 F7HL78 A0A293M4T7 A0A2K6APR7 A0A2K5UNN5 A0A2K5YYA3 A0A2I3MUN9 A0A232FDK6 K7J3J9 A0A2I9LPI3 A0A226F484 I3MFP2 A0A1B6GSF6 R7U9J3 A0A0L8HMP5 A0A1X7UJA5 A7RG79 A0A0K8TPC0 A0A1U7TQF4 A0A1B6MKB1 A0A1B6IDA1 A0A2K6APS0 A0A2K6JUQ8 A0A2K5YY97 A0A0D9QZ02 G7PIG1 A0A2K6QHW7 G7N7A8 A0A384DTH5 D2H337 A0A1A6GXF5 A0A250XWG6 A0A286X8R6 A0A091D0Q5 A0A1S3FN70 B2RPS1 A0A1U7QS77 E2QW69 G1S735 A0A341ATE0 A0A2Y9R4C6 A0A2U3ZYN7 A0A2R9B4V1 A0A2Y9FUJ5 A0A2Y9GU78 A0A2U3XBJ7 A0A2Y9L023 A0A2U3W147 L8Y6K1
D6WJQ8 A0A1W4XB07 N6U3V1 A0A224XXU2 A0A0P4VM87 A0A0V0G6J0 A0A023F9Y3 A0A069DQS6 R4FPV9 A0A023GG88 A0A224Z7I0 A0A023FJI3 L7M4V0 A0A023FV39 A0A131XDE5 A0A1E1X231 A0A1D2N312 E2BP20 A0A1Y1MGW0 V5GWI1 A0A023GF17 A0A2R5L9F5 A0A195E703 F4X6V7 A0A151XC02 A0A2T7PWI0 E9J660 E1ZYZ0 A0A026W7Q2 A0A0J7KXD6 A0A158NFX1 A0A195B2M4 T1J5J1 V5HZ27 A0A2J7Q0P2 A0A067RWS2 V9IG87 K1R1Q8 V3ZPS1 A0A1Z5KWW0 A0A310SMQ2 A0A154PPM4 A0A0L7QJX7 A0A2I3TG75 A0A210QN54 A0A087UC73 F7HL78 A0A293M4T7 A0A2K6APR7 A0A2K5UNN5 A0A2K5YYA3 A0A2I3MUN9 A0A232FDK6 K7J3J9 A0A2I9LPI3 A0A226F484 I3MFP2 A0A1B6GSF6 R7U9J3 A0A0L8HMP5 A0A1X7UJA5 A7RG79 A0A0K8TPC0 A0A1U7TQF4 A0A1B6MKB1 A0A1B6IDA1 A0A2K6APS0 A0A2K6JUQ8 A0A2K5YY97 A0A0D9QZ02 G7PIG1 A0A2K6QHW7 G7N7A8 A0A384DTH5 D2H337 A0A1A6GXF5 A0A250XWG6 A0A286X8R6 A0A091D0Q5 A0A1S3FN70 B2RPS1 A0A1U7QS77 E2QW69 G1S735 A0A341ATE0 A0A2Y9R4C6 A0A2U3ZYN7 A0A2R9B4V1 A0A2Y9FUJ5 A0A2Y9GU78 A0A2U3XBJ7 A0A2Y9L023 A0A2U3W147 L8Y6K1
PDB
2HEI
E-value=1.6246e-82,
Score=776
Ontologies
GO
GO:0003924
GO:0005525
GO:0030100
GO:0032482
GO:0005768
GO:0005769
GO:0030139
GO:0006897
GO:0005886
GO:0006886
GO:0048227
GO:0019003
GO:0070062
GO:0019882
GO:0043231
GO:0007032
GO:0030742
GO:0098993
GO:0070382
GO:0007264
GO:0043066
GO:0043154
GO:0006508
GO:0006811
GO:0016020
GO:0005515
GO:0003779
GO:0016021
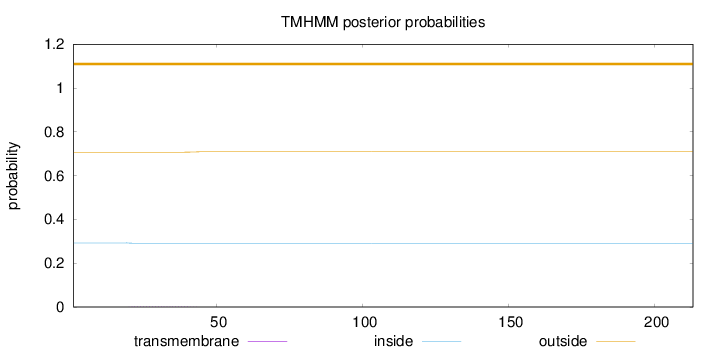
Topology
Length:
213
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0395400000000001
Exp number, first 60 AAs:
0.0362
Total prob of N-in:
0.29279
outside
1 - 213
Population Genetic Test Statistics
Pi
230.283401
Theta
165.751784
Tajima's D
1.166488
CLR
0.507884
CSRT
0.706314684265787
Interpretation
Uncertain
