Gene
KWMTBOMO01659 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006103
Annotation
PREDICTED:_enhancer_of_mRNA-decapping_protein_4-like?_partial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.747
Sequence
CDS
ATGAAAGTGACATTTCTACTAAGGAATTCATGTGGAAAACTGAAAAATAGACGTAAAATGCAACCACCAACCATGTTACCTAAGCTTTCAGATACAACTCAGACGATAGCATTTGCTGAAGGAGATGGTATGAGCTGTATAGAAGTTTACTCTTCGGATGTGGTCGTGACTATCAACACCGGTAACCATGATCATGGTAGCTCAAAAGTGAAGTTGAAGAATCTTGTAGATTATAACTGGGAGCCCAAGTTTTATCCTGGGCAGCTTCTAGCCATTCACATCAGTGGAAAGTACCTGGCATACTCTATTAAAGCTCCTAATACAGCTAACCCAGGTACTTGCAGTGGTATGGTTAGAGTTGTGTACAACCTTGAACCAGGAACTGATCGTCGTGCTCTCATCAAGGGAATGAAGGGGGAGGTCCAAGACTTATCATTTGCCCATATTCAAAACGAGATAGTATTGGCTTGCATCGATGAACAGGGCAACTTTTATGTTCATGAGATTGAAGCAACTGACAGTGGACTGAGATGTACACTAATCGCTGAGATTCGTGAAGATACAGGTATTGTGGGATCATTCCACAGAGTTGTGTGGTGCCCTTATATACCCGATGAAGAAGACGATGACCCTGATGAAGATGTGGATAGACTTCTGCTGACGACTCATGCTAATACTGCCCGCATCTGGAACATGCGCGTGATCGCGAGGGCCGTGGCGGGCACGGAGGGGGTGGGGGGCGCGGGTGCGCTGGCGGGCGGCGCGGGCACGCTGTCCGCGGGCGGGCACGCCGCCGACCTGGTGGACGCCGCCTTCTCGCCCGACGGCACCGCGCTCGCCACCGCCGCCGCCGACGGATACGTCATGTTCTTTCAGGTGTGCATGCAGAACCGGAGCCCCCGCTGCCTGCACAAGTGGCAGCCGCACGACGGGAAGCCCCTCAGCTGTCTCTTCTTCCTCGACAACCACAACAACTACAACTCTGATGTTCAGTTCTGGAAGTTTGCCGTGACCGGCGCGGACAACAACACCTGCATCAAGATATGGTCGTGCAAGTCCTGGAACTGCCTGCAGACCATCTCGTTCGCTCCTGAAGTGCCCAACGAGAGCAGTCTCGGTTTGAAGGTGATGCTGGACAGGAGCGCGCGGTTCCTGGTGGTGTCAGCGGCGAGCGGCCGCGCGCTGTACGTGCTGGGCGTGGCGCGCGACGCGGCCGACACGCACGCACACTGCACCGCGCTCGCGCACTTCCTGCTGCCCTTCCCCGTGCTCAGCTTCTGCATCGCCGACGCGGAAGAACAGACCGCGAAGTGCGAGTCCAGCTGCTCCGACCCGTTCCACCGCAGCGAGTCCGCCGACCCCGCCGGGCCCTCGCCCGACGACTTCGACCTCCAGCCGGACGTGGCGGACTCGGGCTCGGAGTGCGGGTGCGTCGGGCGGGCCGCCTTGCGCCTCTACGTGGTGCAGCCCAAGGGCCTGCAGGAAGGGCGGCTGGCCTTCGCACTGCCTGCAGGCGGCGCGCCCGAGGAGCAGGGCGGCGAGGCCGGGCCGGCGGCCGTGCCCTCCAGCATCCTGCAGCAGCAGAGCCAGCAACTCAAGAACCTGCTCATGAGGTCACAGACGCAGCCCGGGAGCCTCATCGGTCAGAGGGCGGAGTCTCCGCCGATCCCTCCGCAGCTCAACCTCATGACGCCCGACGCGTTCAGCTCGCCGGGCAAGAGGGAGGAAGAGGATCCGCCGCTGTCGGCCACGCCTGACGTCAGCGCCAAGCCGCGCGGCTCCGTCGGAGGGGACGCCGACCGCGCCAAGGGAGCCGAGGGCGCGGGCAAGCCCACCTCGGGGGGGTCCAGTCCCAGCCGGGAGGTGCAGGAGATCATGGCGCACAATGACGACGCCTTCTTCGCCGACAACCTGTGTAGTGTGGGAGCCGCCGACGTCACCAGTTGTTCCGTTACAGACGGCGACACCTCGTGGCCGCAGATATCCATAGCGCAGATAACGGAAGCGAACCAACGCAAGGCGTCCAGCGACAAGTCGGGCGCGCAGAGCCAGCCGCTCGGCGCCGCGTGCGACGCGCGACTCGACCTGCTCGAGCACAAACTGGACAAAATCACAGAGTTGCTGCAGCAGCAGGGTCGCGAGCTGCGGGCCCTGCGCGGCGCCACGTGGTGTCCGCGGACCGCACTGGACGAAGCGCTGCAGGCACACGCACAGGCCTTAGATAATGCTGTCACACTAGGGTGGGAGCGCATGTCGGCGGCGGGCGAGGCTGCGGGCGCGGCGGCGGGCGCGGGCGCGGCGCAGGCGGCGGGCGCGGGGGCGGCGCGCGCGCTGGAGCCGCTGGTGGCCGCGCTGCAGCACGAGCTCGCCGCCAAGCTCACCGCCACCGACCAGCTGCTCAGGGACAACATCGACAAACTAGTCAACAGCAAGGCGGTGTCGGAGCGGCTGAGCTCGTCCATAGCGCGCTGCGTGTCGGAGCAGGTCCGCGAGTCCTTCCGCGCGTCGGTGACGGACAGCCTGCTGCCCGCCATGGAGCGCGCGCACGCACAGATCTTCCGACAAGTCAACAACGCCTTCCAGAACGGGACTAAAGAGTTCGCAGCTCACACGGAGGCCGCAGCCCGCGCGGCGGCGGAGCGCGGCGGGGCGGCGGCCTGCACCGCGCTGCGGGCCGCGCTGGACCGGCACGCCGATACGCTGGCGCAGCACCCGCGCCTGCTGCTGCAGCACGCCACGCAGGCCGTGCGGGACGCGGCGCACAGCGTGCTGGAGAAGGAGCTGCTGTGGTGGCGCGAGCAGGCCCGCTCGGCGTCGCTGACGTCACGCGCGCACTCGCCGCACACGCCGCACGCGCACAACCCGCTCGACCGACAGATGCAAATATCTCAAGTGCAAAACATGATCTCATCGGGTGACGTGAACGGCGCCTTCCAGCTGGCCTTGTCCGCCTCGGACCTGTCCCTCGTGGTCACAGCCTGCAGGTCGGCCGAGCCTTCACTGGTGTTCGGGCCCCCCTGTCAGCTCAAGCAGCACGTACTGCTATCATTAGTTCAGCAACTGGCCGCCGATATGTCCAAGGACACGCAGCTCAAGCACAGGTATCTAGAGGAAGCAGTAATGAACCTGGATACGTCTAGTCCTGTGACCCGTGAGCATCTGCCAGGAGTCGTAGCTGAACTGCAGCAGCAAGTGTCGGCCTTCGTGACCGCCTCGCCCTCGCACCCGCTGGCTCGCAGACTGCGCATGTTGCTGCTGGCCTCTGATGCTCTGCTCAAGGCCACGGTGTGA
Protein
MKVTFLLRNSCGKLKNRRKMQPPTMLPKLSDTTQTIAFAEGDGMSCIEVYSSDVVVTINTGNHDHGSSKVKLKNLVDYNWEPKFYPGQLLAIHISGKYLAYSIKAPNTANPGTCSGMVRVVYNLEPGTDRRALIKGMKGEVQDLSFAHIQNEIVLACIDEQGNFYVHEIEATDSGLRCTLIAEIREDTGIVGSFHRVVWCPYIPDEEDDDPDEDVDRLLLTTHANTARIWNMRVIARAVAGTEGVGGAGALAGGAGTLSAGGHAADLVDAAFSPDGTALATAAADGYVMFFQVCMQNRSPRCLHKWQPHDGKPLSCLFFLDNHNNYNSDVQFWKFAVTGADNNTCIKIWSCKSWNCLQTISFAPEVPNESSLGLKVMLDRSARFLVVSAASGRALYVLGVARDAADTHAHCTALAHFLLPFPVLSFCIADAEEQTAKCESSCSDPFHRSESADPAGPSPDDFDLQPDVADSGSECGCVGRAALRLYVVQPKGLQEGRLAFALPAGGAPEEQGGEAGPAAVPSSILQQQSQQLKNLLMRSQTQPGSLIGQRAESPPIPPQLNLMTPDAFSSPGKREEEDPPLSATPDVSAKPRGSVGGDADRAKGAEGAGKPTSGGSSPSREVQEIMAHNDDAFFADNLCSVGAADVTSCSVTDGDTSWPQISIAQITEANQRKASSDKSGAQSQPLGAACDARLDLLEHKLDKITELLQQQGRELRALRGATWCPRTALDEALQAHAQALDNAVTLGWERMSAAGEAAGAAAGAGAAQAAGAGAARALEPLVAALQHELAAKLTATDQLLRDNIDKLVNSKAVSERLSSSIARCVSEQVRESFRASVTDSLLPAMERAHAQIFRQVNNAFQNGTKEFAAHTEAAARAAAERGGAAACTALRAALDRHADTLAQHPRLLLQHATQAVRDAAHSVLEKELLWWREQARSASLTSRAHSPHTPHAHNPLDRQMQISQVQNMISSGDVNGAFQLALSASDLSLVVTACRSAEPSLVFGPPCQLKQHVLLSLVQQLAADMSKDTQLKHRYLEEAVMNLDTSSPVTREHLPGVVAELQQQVSAFVTASPSHPLARRLRMLLLASDALLKATV
Summary
Uniprot
A0A194PYS7
A0A2A4JUR8
A0A2J7QWF8
A0A0L7RC16
E2AXF8
A0A232EKD6
+ More
A0A1B6MJB4 A0A1Y1L3Y4 A0A154PGB8 A0A310S8S1 F4WSF0 A0A151X3L7 E2BKU7 A0A195ERX8 A0A3L8DDI1 A0A158NTM2 A0A195BRA0 A0A0J7L5J4 A0A026VZ10 A0A023F8T6 A0A224XDY0 A0A069DWY6 A0A0V0G6F1 A0A1B6GNW8 A0A195EFV6 A0A1B6DK13 A0A195D453 A0A1B6EDS9 A0A1B6HFA4 A0A1B0D1F1
A0A1B6MJB4 A0A1Y1L3Y4 A0A154PGB8 A0A310S8S1 F4WSF0 A0A151X3L7 E2BKU7 A0A195ERX8 A0A3L8DDI1 A0A158NTM2 A0A195BRA0 A0A0J7L5J4 A0A026VZ10 A0A023F8T6 A0A224XDY0 A0A069DWY6 A0A0V0G6F1 A0A1B6GNW8 A0A195EFV6 A0A1B6DK13 A0A195D453 A0A1B6EDS9 A0A1B6HFA4 A0A1B0D1F1
Pubmed
EMBL
KQ459584
KPI98492.1
NWSH01000576
PCG75549.1
NEVH01009765
PNF32917.1
+ More
KQ414617 KOC68379.1 GL443549 EFN61882.1 NNAY01003819 OXU18805.1 GEBQ01003951 JAT36026.1 GEZM01065616 JAV68409.1 KQ434899 KZC10882.1 KQ764145 OAD54594.1 GL888321 EGI62867.1 KQ982562 KYQ54850.1 GL448826 EFN83696.1 KQ981993 KYN30983.1 QOIP01000010 RLU17968.1 ADTU01026033 KQ976424 KYM88668.1 LBMM01000489 KMQ98237.1 KK107549 EZA49038.1 GBBI01000895 JAC17817.1 GFTR01008428 JAW07998.1 GBGD01000301 JAC88588.1 GECL01002534 JAP03590.1 GECZ01005660 JAS64109.1 KQ978983 KYN26752.1 GEDC01011306 JAS25992.1 KQ976881 KYN07621.1 GEDC01001202 JAS36096.1 GECU01034299 JAS73407.1 AJVK01010232
KQ414617 KOC68379.1 GL443549 EFN61882.1 NNAY01003819 OXU18805.1 GEBQ01003951 JAT36026.1 GEZM01065616 JAV68409.1 KQ434899 KZC10882.1 KQ764145 OAD54594.1 GL888321 EGI62867.1 KQ982562 KYQ54850.1 GL448826 EFN83696.1 KQ981993 KYN30983.1 QOIP01000010 RLU17968.1 ADTU01026033 KQ976424 KYM88668.1 LBMM01000489 KMQ98237.1 KK107549 EZA49038.1 GBBI01000895 JAC17817.1 GFTR01008428 JAW07998.1 GBGD01000301 JAC88588.1 GECL01002534 JAP03590.1 GECZ01005660 JAS64109.1 KQ978983 KYN26752.1 GEDC01011306 JAS25992.1 KQ976881 KYN07621.1 GEDC01001202 JAS36096.1 GECU01034299 JAS73407.1 AJVK01010232
Proteomes
Pfam
PF16529 Ge1_WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A194PYS7
A0A2A4JUR8
A0A2J7QWF8
A0A0L7RC16
E2AXF8
A0A232EKD6
+ More
A0A1B6MJB4 A0A1Y1L3Y4 A0A154PGB8 A0A310S8S1 F4WSF0 A0A151X3L7 E2BKU7 A0A195ERX8 A0A3L8DDI1 A0A158NTM2 A0A195BRA0 A0A0J7L5J4 A0A026VZ10 A0A023F8T6 A0A224XDY0 A0A069DWY6 A0A0V0G6F1 A0A1B6GNW8 A0A195EFV6 A0A1B6DK13 A0A195D453 A0A1B6EDS9 A0A1B6HFA4 A0A1B0D1F1
A0A1B6MJB4 A0A1Y1L3Y4 A0A154PGB8 A0A310S8S1 F4WSF0 A0A151X3L7 E2BKU7 A0A195ERX8 A0A3L8DDI1 A0A158NTM2 A0A195BRA0 A0A0J7L5J4 A0A026VZ10 A0A023F8T6 A0A224XDY0 A0A069DWY6 A0A0V0G6F1 A0A1B6GNW8 A0A195EFV6 A0A1B6DK13 A0A195D453 A0A1B6EDS9 A0A1B6HFA4 A0A1B0D1F1
PDB
2VXG
E-value=9.70849e-16,
Score=208
Ontologies
GO
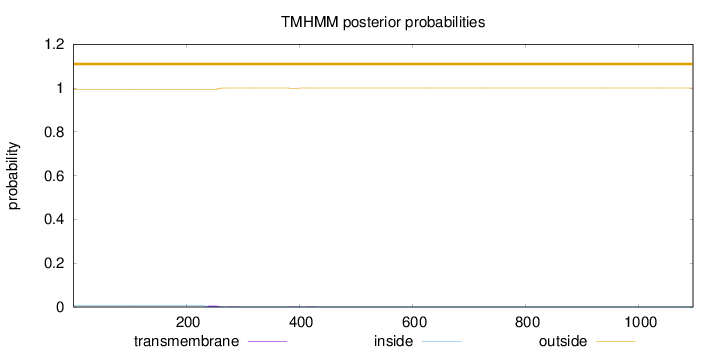
Topology
Length:
1096
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14011
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00557
outside
1 - 1096
Population Genetic Test Statistics
Pi
248.792382
Theta
191.58898
Tajima's D
1.17867
CLR
0.130709
CSRT
0.707014649267537
Interpretation
Uncertain
