Pre Gene Modal
BGIBMGA006047
Annotation
PREDICTED:_calcium_channel_flower_isoform_X1_[Bombyx_mori]
Full name
Calcium channel flower
+ More
Autophagy-related protein 3
Autophagy-related protein 3
Location in the cell
Nuclear Reliability : 2.747
Sequence
CDS
ATGTCGTTAACGGACAAGATATCGATGCTCATGTCGCGTCCGGGGGGTGACAATGCCCAGAAAGATGACGTTCCTACTTGGATGCGATATGCTGGACGGGGTCTTGGAACAGTTGGCAGCTTCATCGCCATCTTCTTTGGTCTCTTCAACTGCTTGGGCATTCTAACCTTGGACGTGCCGTGCCTGATCGCGGGCATATGGCAGATGCTTGCTGGTTTCATTGTCATAGTATGCGAGGCTCCCTGCTGCTGTTTCTTCATCGACTACGTGGGGACCTTGAGCCAGAAAATGGAGACCAGACCTTATTGGAACAAGGCCGCTATGTATCTTGGACTGGCCGTGCCTCCGTTTTTCCTTTGCTTCTTCAGTTTCAGTACGTGGTTGGGTAGCGGACTGATATTCGCCACCGGGATCATATACGGAATGATGGCGCTCGGCAAAAAAGCGTCAATAGAGGACATGCGGTCGTCGGCGGCCAATCTGGAGGCGGGCGTGGGGCCAACAACCGCGCCCCGAGCAACCCTCGTTACCAACGAGCAGCCCGTCAGCTTCACCGGCACGCCTGTCACCAAGTGA
Protein
MSLTDKISMLMSRPGGDNAQKDDVPTWMRYAGRGLGTVGSFIAIFFGLFNCLGILTLDVPCLIAGIWQMLAGFIVIVCEAPCCCFFIDYVGTLSQKMETRPYWNKAAMYLGLAVPPFFLCFFSFSTWLGSGLIFATGIIYGMMALGKKASIEDMRSSAANLEAGVGPTTAPRATLVTNEQPVSFTGTPVTK
Summary
Description
A calcium channel that regulates synaptic endocytosis and hence couples exo- with endocytosis. Required in the nervous system and necessary in photoreceptor cells (By similarity).
A calcium channel that regulates synaptic endocytosis and hence couples exo- with endocytosis. Isoform A and isoform B are mainly required in the nervous system and necessary in photoreceptor cells.
A calcium channel that regulates synaptic endocytosis and hence couples exo- with endocytosis. Isoform A and isoform B are mainly required in the nervous system and necessary in photoreceptor cells.
Subunit
Homomultimer.
Homomultimer; Isoform B.
Homomultimer; Isoform B.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the calcium channel flower family.
Belongs to the ATG3 family.
Belongs to the tetraspanin (TM4SF) family.
Belongs to the calcium channel flower family.
Belongs to the ATG3 family.
Belongs to the tetraspanin (TM4SF) family.
Keywords
Calcium
Calcium channel
Calcium transport
Cell junction
Complete proteome
Cytoplasmic vesicle
Endocytosis
Ion channel
Ion transport
Membrane
Reference proteome
Synapse
Transmembrane
Transmembrane helix
Transport
Alternative splicing
Feature
chain Calcium channel flower
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9J955
A0A194PZM8
A0A1E1WH15
A0A2A4JVY1
S4PFF6
A0A2H1WRF6
+ More
A0A194R7P6 A0A310SD89 A0A0L7R4Q2 A0A0T6AX73 A0A2A3E0W8 A0A1Y1M821 A0A067RNN1 A0A0N0U7H4 A0A026VT25 F4W5W0 E9IJF1 V5GYY9 A0A154P600 A0A158P2U4 A0A2J7RF42 A0A2A3E244 V9IK87 A0A088ADA6 U5EYA5 D6WJ36 A0A0J7N6C2 A0A0C9RCB6 A0A2W1BL35 A0A3L8DS08 A0A1L8DEZ4 A0A1B0CJL6 N6T4S6 A0A195DFH9 A0A195BPM4 A0A182MXU1 A0A069DQG0 A0A224XY48 A0A023EIN2 Q17MR0 A0A0K8TQQ4 A0A182Y5Y6 A0A182G9K9 A0A0P4W0X5 E0VVH2 A0A2M4C0F0 E2AI05 A0A023F010 A0A1B6GRP1 Q7QGH6 A0A182Q7L2 A0A195C2E2 T1E7N1 A0A2M3ZKK9 A0A2M4AMR1 A0A182UV36 A0A182WEU8 A0A0V0G7P7 A0A182MHX3 A0A195EYK5 T1HDB7 A0A182LP65 A0A182U8J1 A0A182HNB2 A0A182XC14 A0A0A9X349 E2BZ86 A0A182RD38 W5J8H2 A0A087ZIR7 A0A1Q3FY30 Q17MQ9 B3M9W1 Q17MQ8 A0A1W7R5A9 A0A1W4WHP6 A0A0M3QWB9 A0A1Q3G3K1 A0A182FBI5 A0A232EN66 B4J043 A0A1W4VNR7 A0A3B0KIX6 A0A0A1XQP6 A0A212EZ16 A0A1B6FQY0 Q95T12 A0A182JIK1 A0A2M4AQT0 A0A1B6CZ35 A0A0J9ULS9 B4QLP9 A0A0R3P864 B4GRI8 B4PD01 A0A0A9WJ01 A0A1A9V4E8 A0A182PKW0 D3TQ18 B3NDM7
A0A194R7P6 A0A310SD89 A0A0L7R4Q2 A0A0T6AX73 A0A2A3E0W8 A0A1Y1M821 A0A067RNN1 A0A0N0U7H4 A0A026VT25 F4W5W0 E9IJF1 V5GYY9 A0A154P600 A0A158P2U4 A0A2J7RF42 A0A2A3E244 V9IK87 A0A088ADA6 U5EYA5 D6WJ36 A0A0J7N6C2 A0A0C9RCB6 A0A2W1BL35 A0A3L8DS08 A0A1L8DEZ4 A0A1B0CJL6 N6T4S6 A0A195DFH9 A0A195BPM4 A0A182MXU1 A0A069DQG0 A0A224XY48 A0A023EIN2 Q17MR0 A0A0K8TQQ4 A0A182Y5Y6 A0A182G9K9 A0A0P4W0X5 E0VVH2 A0A2M4C0F0 E2AI05 A0A023F010 A0A1B6GRP1 Q7QGH6 A0A182Q7L2 A0A195C2E2 T1E7N1 A0A2M3ZKK9 A0A2M4AMR1 A0A182UV36 A0A182WEU8 A0A0V0G7P7 A0A182MHX3 A0A195EYK5 T1HDB7 A0A182LP65 A0A182U8J1 A0A182HNB2 A0A182XC14 A0A0A9X349 E2BZ86 A0A182RD38 W5J8H2 A0A087ZIR7 A0A1Q3FY30 Q17MQ9 B3M9W1 Q17MQ8 A0A1W7R5A9 A0A1W4WHP6 A0A0M3QWB9 A0A1Q3G3K1 A0A182FBI5 A0A232EN66 B4J043 A0A1W4VNR7 A0A3B0KIX6 A0A0A1XQP6 A0A212EZ16 A0A1B6FQY0 Q95T12 A0A182JIK1 A0A2M4AQT0 A0A1B6CZ35 A0A0J9ULS9 B4QLP9 A0A0R3P864 B4GRI8 B4PD01 A0A0A9WJ01 A0A1A9V4E8 A0A182PKW0 D3TQ18 B3NDM7
Pubmed
19121390
26354079
23622113
28004739
24845553
24508170
+ More
21719571 21282665 21347285 18362917 19820115 28756777 30249741 23537049 26334808 24945155 17510324 26369729 25244985 26483478 27129103 20566863 20798317 25474469 12364791 14747013 17210077 20966253 25401762 26823975 20920257 17994087 28648823 25830018 22118469 10731132 12537572 12537569 19737521 22936249 15632085 23185243 20353571
21719571 21282665 21347285 18362917 19820115 28756777 30249741 23537049 26334808 24945155 17510324 26369729 25244985 26483478 27129103 20566863 20798317 25474469 12364791 14747013 17210077 20966253 25401762 26823975 20920257 17994087 28648823 25830018 22118469 10731132 12537572 12537569 19737521 22936249 15632085 23185243 20353571
EMBL
BABH01004402
KQ459584
KPI98493.1
GDQN01004903
JAT86151.1
NWSH01000576
+ More
PCG75550.1 GAIX01006610 JAA85950.1 ODYU01010496 SOQ55643.1 KQ460644 KPJ13260.1 KQ762903 OAD55294.1 KQ414657 KOC65845.1 LJIG01022598 KRT79722.1 KZ288467 PBC25403.1 GEZM01038240 JAV81701.1 KK852514 KDR22200.1 KQ435698 KOX80744.1 KK108116 EZA46892.1 GL887695 EGI70436.1 GL763766 EFZ19301.1 GALX01001364 JAB67102.1 KQ434823 KZC07297.1 ADTU01007483 NEVH01004414 PNF39437.1 PBC25402.1 JR049712 AEY61047.1 GANO01000482 JAB59389.1 KQ971342 EFA03876.1 LBMM01009288 KMQ88245.1 GBYB01005950 JAG75717.1 KZ150105 PZC73440.1 QOIP01000005 RLU23096.1 GFDF01009052 JAV05032.1 AJWK01014723 AJWK01014724 APGK01044152 KB741020 KB631628 ENN75139.1 ERL84853.1 KQ980903 KYN11638.1 KQ976432 KYM87447.1 GBGD01002998 JAC85891.1 GFTR01003293 JAW13133.1 GAPW01004300 JAC09298.1 CH477204 EAT47962.1 GDAI01000919 JAI16684.1 JXUM01049369 JXUM01049370 JXUM01049371 KQ561601 KXJ78031.1 GDKW01000050 JAI56545.1 DS235811 EEB17378.1 GGFJ01009317 MBW58458.1 GL439621 EFN66913.1 GBBI01003865 JAC14847.1 GECZ01019535 GECZ01004663 JAS50234.1 JAS65106.1 AAAB01008834 EAA05716.4 AXCN02002117 KQ978344 KYM94992.1 GAMD01002887 JAA98703.1 GGFM01008313 MBW29064.1 GGFK01008716 MBW42037.1 GECL01002104 JAP04020.1 AXCM01005743 KQ981905 KYN33375.1 ACPB03015513 ACPB03015514 ACPB03015515 APCN01001900 GBHO01029175 GBRD01011351 GDHC01017176 JAG14429.1 JAG54473.1 JAQ01453.1 GL451577 EFN78992.1 ADMH02002103 ETN59089.1 GGFL01004627 MBW68805.1 GFDL01002575 JAV32470.1 EAT47963.1 CH902618 EDV40152.1 EAT47964.1 GEHC01001277 JAV46368.1 CP012525 ALC43873.1 GFDL01000660 JAV34385.1 NNAY01003207 OXU19790.1 CH916366 EDV97836.1 OUUW01000009 SPP85041.1 GBXI01000976 JAD13316.1 AGBW02011391 OWR46736.1 GECZ01018114 GECZ01017171 JAS51655.1 JAS52598.1 AE014296 AY060385 AAL25424.1 AAN11767.1 GGFK01009637 MBW42958.1 GEDC01026658 GEDC01026102 GEDC01018673 GEDC01002784 JAS10640.1 JAS11196.1 JAS18625.1 JAS34514.1 CM002912 KMY99825.1 CM000363 EDX10598.1 CH379069 KRT07766.1 KRT07767.1 CH479188 EDW40373.1 CM000159 EDW94933.1 GBHO01038769 GDHC01000405 JAG04835.1 JAQ18224.1 EZ423520 ADD19796.1 CH954178 EDV52160.1
PCG75550.1 GAIX01006610 JAA85950.1 ODYU01010496 SOQ55643.1 KQ460644 KPJ13260.1 KQ762903 OAD55294.1 KQ414657 KOC65845.1 LJIG01022598 KRT79722.1 KZ288467 PBC25403.1 GEZM01038240 JAV81701.1 KK852514 KDR22200.1 KQ435698 KOX80744.1 KK108116 EZA46892.1 GL887695 EGI70436.1 GL763766 EFZ19301.1 GALX01001364 JAB67102.1 KQ434823 KZC07297.1 ADTU01007483 NEVH01004414 PNF39437.1 PBC25402.1 JR049712 AEY61047.1 GANO01000482 JAB59389.1 KQ971342 EFA03876.1 LBMM01009288 KMQ88245.1 GBYB01005950 JAG75717.1 KZ150105 PZC73440.1 QOIP01000005 RLU23096.1 GFDF01009052 JAV05032.1 AJWK01014723 AJWK01014724 APGK01044152 KB741020 KB631628 ENN75139.1 ERL84853.1 KQ980903 KYN11638.1 KQ976432 KYM87447.1 GBGD01002998 JAC85891.1 GFTR01003293 JAW13133.1 GAPW01004300 JAC09298.1 CH477204 EAT47962.1 GDAI01000919 JAI16684.1 JXUM01049369 JXUM01049370 JXUM01049371 KQ561601 KXJ78031.1 GDKW01000050 JAI56545.1 DS235811 EEB17378.1 GGFJ01009317 MBW58458.1 GL439621 EFN66913.1 GBBI01003865 JAC14847.1 GECZ01019535 GECZ01004663 JAS50234.1 JAS65106.1 AAAB01008834 EAA05716.4 AXCN02002117 KQ978344 KYM94992.1 GAMD01002887 JAA98703.1 GGFM01008313 MBW29064.1 GGFK01008716 MBW42037.1 GECL01002104 JAP04020.1 AXCM01005743 KQ981905 KYN33375.1 ACPB03015513 ACPB03015514 ACPB03015515 APCN01001900 GBHO01029175 GBRD01011351 GDHC01017176 JAG14429.1 JAG54473.1 JAQ01453.1 GL451577 EFN78992.1 ADMH02002103 ETN59089.1 GGFL01004627 MBW68805.1 GFDL01002575 JAV32470.1 EAT47963.1 CH902618 EDV40152.1 EAT47964.1 GEHC01001277 JAV46368.1 CP012525 ALC43873.1 GFDL01000660 JAV34385.1 NNAY01003207 OXU19790.1 CH916366 EDV97836.1 OUUW01000009 SPP85041.1 GBXI01000976 JAD13316.1 AGBW02011391 OWR46736.1 GECZ01018114 GECZ01017171 JAS51655.1 JAS52598.1 AE014296 AY060385 AAL25424.1 AAN11767.1 GGFK01009637 MBW42958.1 GEDC01026658 GEDC01026102 GEDC01018673 GEDC01002784 JAS10640.1 JAS11196.1 JAS18625.1 JAS34514.1 CM002912 KMY99825.1 CM000363 EDX10598.1 CH379069 KRT07766.1 KRT07767.1 CH479188 EDW40373.1 CM000159 EDW94933.1 GBHO01038769 GDHC01000405 JAG04835.1 JAQ18224.1 EZ423520 ADD19796.1 CH954178 EDV52160.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000053825
UP000242457
+ More
UP000027135 UP000053105 UP000053097 UP000007755 UP000076502 UP000005205 UP000235965 UP000005203 UP000007266 UP000036403 UP000279307 UP000092461 UP000019118 UP000030742 UP000078492 UP000078540 UP000075884 UP000008820 UP000076408 UP000069940 UP000249989 UP000009046 UP000000311 UP000007062 UP000075886 UP000078542 UP000075903 UP000075920 UP000075883 UP000078541 UP000015103 UP000075882 UP000075902 UP000075840 UP000076407 UP000008237 UP000075900 UP000000673 UP000007801 UP000192223 UP000092553 UP000069272 UP000215335 UP000001070 UP000192221 UP000268350 UP000007151 UP000000803 UP000075880 UP000000304 UP000001819 UP000008744 UP000002282 UP000078200 UP000075885 UP000008711
UP000027135 UP000053105 UP000053097 UP000007755 UP000076502 UP000005205 UP000235965 UP000005203 UP000007266 UP000036403 UP000279307 UP000092461 UP000019118 UP000030742 UP000078492 UP000078540 UP000075884 UP000008820 UP000076408 UP000069940 UP000249989 UP000009046 UP000000311 UP000007062 UP000075886 UP000078542 UP000075903 UP000075920 UP000075883 UP000078541 UP000015103 UP000075882 UP000075902 UP000075840 UP000076407 UP000008237 UP000075900 UP000000673 UP000007801 UP000192223 UP000092553 UP000069272 UP000215335 UP000001070 UP000192221 UP000268350 UP000007151 UP000000803 UP000075880 UP000000304 UP000001819 UP000008744 UP000002282 UP000078200 UP000075885 UP000008711
Pfam
Interpro
IPR019365
TVP18/Ca-channel_flower
+ More
IPR036961 Kinesin_motor_dom_sf
IPR019821 Kinesin_motor_CS
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR019461 Autophagy-rel_prot_3_C
IPR008952 Tetraspanin_EC2_sf
IPR007134 Autophagy-rel_prot_3_N
IPR007135 Autophagy-rel_prot_3
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR013087 Znf_C2H2_type
IPR015318 Znf_GAGA-bd_fac
IPR018499 Tetraspanin/Peripherin
IPR036961 Kinesin_motor_dom_sf
IPR019821 Kinesin_motor_CS
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR019461 Autophagy-rel_prot_3_C
IPR008952 Tetraspanin_EC2_sf
IPR007134 Autophagy-rel_prot_3_N
IPR007135 Autophagy-rel_prot_3
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR013087 Znf_C2H2_type
IPR015318 Znf_GAGA-bd_fac
IPR018499 Tetraspanin/Peripherin
Gene 3D
CDD
ProteinModelPortal
H9J955
A0A194PZM8
A0A1E1WH15
A0A2A4JVY1
S4PFF6
A0A2H1WRF6
+ More
A0A194R7P6 A0A310SD89 A0A0L7R4Q2 A0A0T6AX73 A0A2A3E0W8 A0A1Y1M821 A0A067RNN1 A0A0N0U7H4 A0A026VT25 F4W5W0 E9IJF1 V5GYY9 A0A154P600 A0A158P2U4 A0A2J7RF42 A0A2A3E244 V9IK87 A0A088ADA6 U5EYA5 D6WJ36 A0A0J7N6C2 A0A0C9RCB6 A0A2W1BL35 A0A3L8DS08 A0A1L8DEZ4 A0A1B0CJL6 N6T4S6 A0A195DFH9 A0A195BPM4 A0A182MXU1 A0A069DQG0 A0A224XY48 A0A023EIN2 Q17MR0 A0A0K8TQQ4 A0A182Y5Y6 A0A182G9K9 A0A0P4W0X5 E0VVH2 A0A2M4C0F0 E2AI05 A0A023F010 A0A1B6GRP1 Q7QGH6 A0A182Q7L2 A0A195C2E2 T1E7N1 A0A2M3ZKK9 A0A2M4AMR1 A0A182UV36 A0A182WEU8 A0A0V0G7P7 A0A182MHX3 A0A195EYK5 T1HDB7 A0A182LP65 A0A182U8J1 A0A182HNB2 A0A182XC14 A0A0A9X349 E2BZ86 A0A182RD38 W5J8H2 A0A087ZIR7 A0A1Q3FY30 Q17MQ9 B3M9W1 Q17MQ8 A0A1W7R5A9 A0A1W4WHP6 A0A0M3QWB9 A0A1Q3G3K1 A0A182FBI5 A0A232EN66 B4J043 A0A1W4VNR7 A0A3B0KIX6 A0A0A1XQP6 A0A212EZ16 A0A1B6FQY0 Q95T12 A0A182JIK1 A0A2M4AQT0 A0A1B6CZ35 A0A0J9ULS9 B4QLP9 A0A0R3P864 B4GRI8 B4PD01 A0A0A9WJ01 A0A1A9V4E8 A0A182PKW0 D3TQ18 B3NDM7
A0A194R7P6 A0A310SD89 A0A0L7R4Q2 A0A0T6AX73 A0A2A3E0W8 A0A1Y1M821 A0A067RNN1 A0A0N0U7H4 A0A026VT25 F4W5W0 E9IJF1 V5GYY9 A0A154P600 A0A158P2U4 A0A2J7RF42 A0A2A3E244 V9IK87 A0A088ADA6 U5EYA5 D6WJ36 A0A0J7N6C2 A0A0C9RCB6 A0A2W1BL35 A0A3L8DS08 A0A1L8DEZ4 A0A1B0CJL6 N6T4S6 A0A195DFH9 A0A195BPM4 A0A182MXU1 A0A069DQG0 A0A224XY48 A0A023EIN2 Q17MR0 A0A0K8TQQ4 A0A182Y5Y6 A0A182G9K9 A0A0P4W0X5 E0VVH2 A0A2M4C0F0 E2AI05 A0A023F010 A0A1B6GRP1 Q7QGH6 A0A182Q7L2 A0A195C2E2 T1E7N1 A0A2M3ZKK9 A0A2M4AMR1 A0A182UV36 A0A182WEU8 A0A0V0G7P7 A0A182MHX3 A0A195EYK5 T1HDB7 A0A182LP65 A0A182U8J1 A0A182HNB2 A0A182XC14 A0A0A9X349 E2BZ86 A0A182RD38 W5J8H2 A0A087ZIR7 A0A1Q3FY30 Q17MQ9 B3M9W1 Q17MQ8 A0A1W7R5A9 A0A1W4WHP6 A0A0M3QWB9 A0A1Q3G3K1 A0A182FBI5 A0A232EN66 B4J043 A0A1W4VNR7 A0A3B0KIX6 A0A0A1XQP6 A0A212EZ16 A0A1B6FQY0 Q95T12 A0A182JIK1 A0A2M4AQT0 A0A1B6CZ35 A0A0J9ULS9 B4QLP9 A0A0R3P864 B4GRI8 B4PD01 A0A0A9WJ01 A0A1A9V4E8 A0A182PKW0 D3TQ18 B3NDM7
Ontologies
GO
GO:0016021
GO:0016192
GO:0008017
GO:0005524
GO:0007018
GO:0003777
GO:0003676
GO:0043525
GO:0150008
GO:0099533
GO:0048488
GO:0150007
GO:0044214
GO:0030285
GO:0005262
GO:0042802
GO:0046530
GO:0030054
GO:0035212
GO:0005737
GO:0015031
GO:0006914
GO:0042981
GO:0070588
GO:0006816
GO:0005886
GO:0019079
GO:0043066
GO:0043154
GO:0006508
GO:0006811
GO:0016020
GO:0005515
GO:0003779
Topology
Subcellular location
Upon fusion of the synaptic vesicle with the presynaptic membrane, protein is present in the periactive zones, where endocytosis is known to occur. With evidence from 1 publications.
Cytoplasm
Cytoplasm
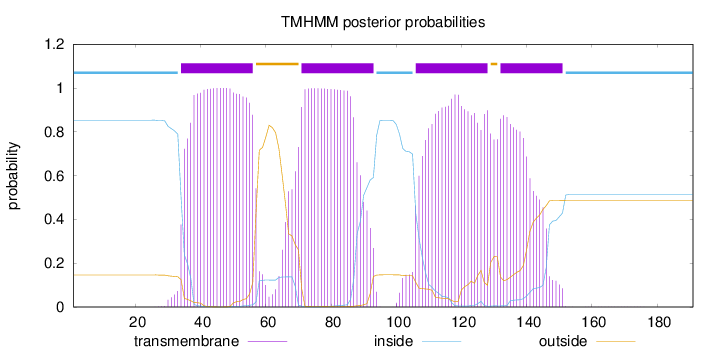
Length:
191
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
78.67001
Exp number, first 60 AAs:
22.43114
Total prob of N-in:
0.85334
POSSIBLE N-term signal
sequence
inside
1 - 33
TMhelix
34 - 56
outside
57 - 70
TMhelix
71 - 93
inside
94 - 105
TMhelix
106 - 128
outside
129 - 131
TMhelix
132 - 151
inside
152 - 191
Population Genetic Test Statistics
Pi
165.171363
Theta
156.158753
Tajima's D
-0.626171
CLR
10.40874
CSRT
0.210139493025349
Interpretation
Uncertain
