Gene
KWMTBOMO01655
Pre Gene Modal
BGIBMGA006048
Annotation
PREDICTED:_inactive_hydroxysteroid_dehydrogenase-like_protein_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.98
Sequence
CDS
ATGTTTTTCACGTTGTTAATAGTTCTCCTAGTTGTAATCGGTTTGTTGGCTGTGTTGTCGATTGCTCTGGACTCGCTGTGGAGTCCTTTAGAGTTGGTTTCGTCTTATTTGATGCCGTACTTCCTGCCCGCCGAGGATCAGCCGCTGACCAAAAAGTTCGGATCGTGGGCCGTTGTGACCGGCGGCACGGACGGTATCGGTAAGCACTACGCCAAGGAGCTGGCACGACGCGGCCTCAACGTGGTCATCATCAGCAGAAACCCTGAAAAATTGAAAGCAACGGTCACTGAAATCGAAAAAGAACACAACGTGAAAACCAAGATGGTGGTTGCTGACTTCAGTAAAGGCCTTGAAGTGTACGCTCATATCGAAGAGGAACTCAAAGACATTCCTATCGGTATACTAGTGAACAATGTTGGCGTACAGTACGATTACCCGACGGAGCTTGGTGACCTGCCGATGTCGAAAGCGTGGGAGATAGTCCACGTGAATGTCGGAGCCGTTACCAGTATGACCAGGCTCGTGGTAAAGGAGATGGCCAAGAGGGGAAAGGGTGCTGTGGTCAACGTGTCTTCCGGCTCCGAGCTGCAGCCACTGCCGTTGATGGCTGTGTACGCAGCTACCAAGGCTTACGTTCGCAGTTTCACACTGGCGATTCGCGAAGAATACGCTCCTAAAGGGATCCACGTTCAGCACATGTCGCCACTTTTCGTGTCAACGAAGATGAACTCGTTCTCCCAGCGGCTTCTCGATGGGAACCTGCTGGTGCCTGATGCTGCCACTTACGCAAAACACGCAGCGGCTACTCTCGGTCGTGTACATGACACCACTGGGTATTGGCTTCATGGAATACAGGTATGGACGTAA
Protein
MFFTLLIVLLVVIGLLAVLSIALDSLWSPLELVSSYLMPYFLPAEDQPLTKKFGSWAVVTGGTDGIGKHYAKELARRGLNVVIISRNPEKLKATVTEIEKEHNVKTKMVVADFSKGLEVYAHIEEELKDIPIGILVNNVGVQYDYPTELGDLPMSKAWEIVHVNVGAVTSMTRLVVKEMAKRGKGAVVNVSSGSELQPLPLMAVYAATKAYVRSFTLAIREEYAPKGIHVQHMSPLFVSTKMNSFSQRLLDGNLLVPDAATYAKHAAATLGRVHDTTGYWLHGIQVWT
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J956
A0A2W1BF62
A0A194Q0G6
I4DPM9
A0A194R616
A0A1L8DZ69
+ More
A0A1L8DYP3 A0A1B0GKX7 A0A1Y1M4L3 A0A2A4JG60 D6WN07 A0A1Y1M9I6 A0A0L0BS07 A0A1A9WL20 A0A2H1W2C5 W8BCS0 W8BHQ7 A0A0A1WFH5 A0A0A1WT83 A0A2C9GS57 A0A182MP58 A0A023F3V8 K7J1P8 A0A224XKX2 A0A1S4GYN2 A0A182UUY6 A0A182KMI3 A0A182UG21 A0A182IQG5 A0A0V0G9A4 A0A0K8VLN1 A0A182NTA5 A0A084VUV2 A0A182QW36 A0A0P6JSU2 A0A1I8ML52 A0A023ENN4 W5J4S0 A0A182P4K2 A0A0K8UZP2 T1PJR0 A0A182F1A4 A0A212EMX8 A0A182RWA4 A0A182Y9P5 A0A034W898 U4UJN8 A0A1A9WL19 A0A1I8NPE7 N6ULG0 J3JUC0 B0WNB3 A0A1A9XT88 A0A2S2NBB4 A0A0L7RI89 A0A182WRV7 A0A1A9ZCY6 A0A1Q3F1E5 C4WU15 A0A023EPR8 A0A232F1P4 A0A1B0FG51 A0A0A9YJR7 A0A1B6EBX6 A0A1B6L3U3 A0A1B6IV31 A0A2H8TMP7 T1HFB4 U5EIA3 A0A1L8DYQ4 A0A1J1HF19 A0A2S2QSF1 A0A2J7QJW4 A0A2A3E1T5 A0A0A1XRZ1 A0A0Q9XD10 B4KJQ9 A0A0M4E2G4 A0A3L8DPZ3 A0A026VVA5 A0A088AAE3 A0A195CJW5 A0A151X1F4 A0A195ETR6 A0A195DVJ3 F4W3Z5 B4LV86 E2APP5 B4JQW7 T1DPK7 A0A0J7NWS7 E2BBS8 A0A1A9UVE8 B4I563 A0A1A9WL14 B4N915 A0A336KUV6 A0A1B0AUP3 A0A1W4VXF8 A0A1W4VWK6
A0A1L8DYP3 A0A1B0GKX7 A0A1Y1M4L3 A0A2A4JG60 D6WN07 A0A1Y1M9I6 A0A0L0BS07 A0A1A9WL20 A0A2H1W2C5 W8BCS0 W8BHQ7 A0A0A1WFH5 A0A0A1WT83 A0A2C9GS57 A0A182MP58 A0A023F3V8 K7J1P8 A0A224XKX2 A0A1S4GYN2 A0A182UUY6 A0A182KMI3 A0A182UG21 A0A182IQG5 A0A0V0G9A4 A0A0K8VLN1 A0A182NTA5 A0A084VUV2 A0A182QW36 A0A0P6JSU2 A0A1I8ML52 A0A023ENN4 W5J4S0 A0A182P4K2 A0A0K8UZP2 T1PJR0 A0A182F1A4 A0A212EMX8 A0A182RWA4 A0A182Y9P5 A0A034W898 U4UJN8 A0A1A9WL19 A0A1I8NPE7 N6ULG0 J3JUC0 B0WNB3 A0A1A9XT88 A0A2S2NBB4 A0A0L7RI89 A0A182WRV7 A0A1A9ZCY6 A0A1Q3F1E5 C4WU15 A0A023EPR8 A0A232F1P4 A0A1B0FG51 A0A0A9YJR7 A0A1B6EBX6 A0A1B6L3U3 A0A1B6IV31 A0A2H8TMP7 T1HFB4 U5EIA3 A0A1L8DYQ4 A0A1J1HF19 A0A2S2QSF1 A0A2J7QJW4 A0A2A3E1T5 A0A0A1XRZ1 A0A0Q9XD10 B4KJQ9 A0A0M4E2G4 A0A3L8DPZ3 A0A026VVA5 A0A088AAE3 A0A195CJW5 A0A151X1F4 A0A195ETR6 A0A195DVJ3 F4W3Z5 B4LV86 E2APP5 B4JQW7 T1DPK7 A0A0J7NWS7 E2BBS8 A0A1A9UVE8 B4I563 A0A1A9WL14 B4N915 A0A336KUV6 A0A1B0AUP3 A0A1W4VXF8 A0A1W4VWK6
Pubmed
EMBL
BABH01004399
KZ150105
PZC73438.1
KQ459584
KPI98489.1
AK403684
+ More
BAM19869.1 KQ460644 KPJ13263.1 GFDF01002502 JAV11582.1 GFDF01002501 JAV11583.1 AJWK01033922 AJWK01033923 AJWK01033924 GEZM01042277 JAV79978.1 NWSH01001696 PCG70403.1 KQ971343 EFA04333.2 GEZM01042278 JAV79977.1 JRES01001454 KNC22806.1 ODYU01005870 SOQ47198.1 GAMC01010098 JAB96457.1 GAMC01010097 JAB96458.1 GBXI01016861 GBXI01006921 JAC97430.1 JAD07371.1 GBXI01012481 JAD01811.1 APCN01000722 AXCM01001453 AXCM01001454 AXCM01001455 GBBI01002616 JAC16096.1 GFTR01004702 JAW11724.1 AAAB01008944 GECL01001534 JAP04590.1 GDHF01012523 JAI39791.1 ATLV01017026 KE525143 KFB41746.1 AXCN02000718 GDUN01000001 JAN95918.1 GAPW01002566 JAC11032.1 ADMH02002200 ETN57860.1 GDHF01032144 GDHF01020172 GDHF01012506 GDHF01002857 GDHF01000530 JAI20170.1 JAI32142.1 JAI39808.1 JAI49457.1 JAI51784.1 KA648188 AFP62817.1 AGBW02013781 OWR42829.1 GAKP01008018 JAC50934.1 KB632224 ERL90215.1 APGK01028290 APGK01028291 APGK01028292 KB740686 ENN79537.1 BT126833 AEE61795.1 DS232009 EDS31561.1 GGMR01001633 MBY14252.1 KQ414585 KOC70544.1 GFDL01013737 JAV21308.1 ABLF02034255 ABLF02034261 AK340863 BAH71385.1 GAPW01002565 JAC11033.1 NNAY01001239 OXU24641.1 CCAG010010519 GBHO01012261 JAG31343.1 GEDC01001908 JAS35390.1 GEBQ01021642 JAT18335.1 GECU01016909 JAS90797.1 GFXV01003127 MBW14932.1 ACPB03015453 ACPB03015454 GANO01002709 JAB57162.1 GFDF01002527 JAV11557.1 CVRI01000001 CRK86576.1 GGMS01011441 MBY80644.1 NEVH01013548 PNF28881.1 KZ288444 PBC25660.1 GBXI01000545 JAD13747.1 CH933807 KRG02724.1 EDW11504.1 CP012523 ALC39791.1 QOIP01000005 RLU22467.1 KK107847 EZA47466.1 KQ977642 KYN01020.1 KQ982585 KYQ54227.1 KQ981979 KYN31551.1 KQ980295 KYN16898.1 GL887491 EGI70991.1 CH940649 EDW64346.2 GL441572 EFN64596.1 CH916372 EDV99297.1 GAMD01001845 JAA99745.1 LBMM01001129 KMQ96870.1 GL447151 EFN86854.1 CH480822 EDW55519.1 CH964232 EDW80520.1 UFQS01000988 UFQT01000988 SSX08266.1 SSX28309.1 JXJN01003706
BAM19869.1 KQ460644 KPJ13263.1 GFDF01002502 JAV11582.1 GFDF01002501 JAV11583.1 AJWK01033922 AJWK01033923 AJWK01033924 GEZM01042277 JAV79978.1 NWSH01001696 PCG70403.1 KQ971343 EFA04333.2 GEZM01042278 JAV79977.1 JRES01001454 KNC22806.1 ODYU01005870 SOQ47198.1 GAMC01010098 JAB96457.1 GAMC01010097 JAB96458.1 GBXI01016861 GBXI01006921 JAC97430.1 JAD07371.1 GBXI01012481 JAD01811.1 APCN01000722 AXCM01001453 AXCM01001454 AXCM01001455 GBBI01002616 JAC16096.1 GFTR01004702 JAW11724.1 AAAB01008944 GECL01001534 JAP04590.1 GDHF01012523 JAI39791.1 ATLV01017026 KE525143 KFB41746.1 AXCN02000718 GDUN01000001 JAN95918.1 GAPW01002566 JAC11032.1 ADMH02002200 ETN57860.1 GDHF01032144 GDHF01020172 GDHF01012506 GDHF01002857 GDHF01000530 JAI20170.1 JAI32142.1 JAI39808.1 JAI49457.1 JAI51784.1 KA648188 AFP62817.1 AGBW02013781 OWR42829.1 GAKP01008018 JAC50934.1 KB632224 ERL90215.1 APGK01028290 APGK01028291 APGK01028292 KB740686 ENN79537.1 BT126833 AEE61795.1 DS232009 EDS31561.1 GGMR01001633 MBY14252.1 KQ414585 KOC70544.1 GFDL01013737 JAV21308.1 ABLF02034255 ABLF02034261 AK340863 BAH71385.1 GAPW01002565 JAC11033.1 NNAY01001239 OXU24641.1 CCAG010010519 GBHO01012261 JAG31343.1 GEDC01001908 JAS35390.1 GEBQ01021642 JAT18335.1 GECU01016909 JAS90797.1 GFXV01003127 MBW14932.1 ACPB03015453 ACPB03015454 GANO01002709 JAB57162.1 GFDF01002527 JAV11557.1 CVRI01000001 CRK86576.1 GGMS01011441 MBY80644.1 NEVH01013548 PNF28881.1 KZ288444 PBC25660.1 GBXI01000545 JAD13747.1 CH933807 KRG02724.1 EDW11504.1 CP012523 ALC39791.1 QOIP01000005 RLU22467.1 KK107847 EZA47466.1 KQ977642 KYN01020.1 KQ982585 KYQ54227.1 KQ981979 KYN31551.1 KQ980295 KYN16898.1 GL887491 EGI70991.1 CH940649 EDW64346.2 GL441572 EFN64596.1 CH916372 EDV99297.1 GAMD01001845 JAA99745.1 LBMM01001129 KMQ96870.1 GL447151 EFN86854.1 CH480822 EDW55519.1 CH964232 EDW80520.1 UFQS01000988 UFQT01000988 SSX08266.1 SSX28309.1 JXJN01003706
Proteomes
UP000005204
UP000053268
UP000053240
UP000092461
UP000218220
UP000007266
+ More
UP000037069 UP000091820 UP000075840 UP000075883 UP000002358 UP000075903 UP000075882 UP000075902 UP000075880 UP000075884 UP000030765 UP000075886 UP000095301 UP000000673 UP000075885 UP000069272 UP000007151 UP000075900 UP000076408 UP000030742 UP000095300 UP000019118 UP000002320 UP000092443 UP000053825 UP000076407 UP000092445 UP000007819 UP000215335 UP000092444 UP000015103 UP000183832 UP000235965 UP000242457 UP000009192 UP000092553 UP000279307 UP000053097 UP000005203 UP000078542 UP000075809 UP000078541 UP000078492 UP000007755 UP000008792 UP000000311 UP000001070 UP000036403 UP000008237 UP000078200 UP000001292 UP000007798 UP000092460 UP000192221
UP000037069 UP000091820 UP000075840 UP000075883 UP000002358 UP000075903 UP000075882 UP000075902 UP000075880 UP000075884 UP000030765 UP000075886 UP000095301 UP000000673 UP000075885 UP000069272 UP000007151 UP000075900 UP000076408 UP000030742 UP000095300 UP000019118 UP000002320 UP000092443 UP000053825 UP000076407 UP000092445 UP000007819 UP000215335 UP000092444 UP000015103 UP000183832 UP000235965 UP000242457 UP000009192 UP000092553 UP000279307 UP000053097 UP000005203 UP000078542 UP000075809 UP000078541 UP000078492 UP000007755 UP000008792 UP000000311 UP000001070 UP000036403 UP000008237 UP000078200 UP000001292 UP000007798 UP000092460 UP000192221
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J956
A0A2W1BF62
A0A194Q0G6
I4DPM9
A0A194R616
A0A1L8DZ69
+ More
A0A1L8DYP3 A0A1B0GKX7 A0A1Y1M4L3 A0A2A4JG60 D6WN07 A0A1Y1M9I6 A0A0L0BS07 A0A1A9WL20 A0A2H1W2C5 W8BCS0 W8BHQ7 A0A0A1WFH5 A0A0A1WT83 A0A2C9GS57 A0A182MP58 A0A023F3V8 K7J1P8 A0A224XKX2 A0A1S4GYN2 A0A182UUY6 A0A182KMI3 A0A182UG21 A0A182IQG5 A0A0V0G9A4 A0A0K8VLN1 A0A182NTA5 A0A084VUV2 A0A182QW36 A0A0P6JSU2 A0A1I8ML52 A0A023ENN4 W5J4S0 A0A182P4K2 A0A0K8UZP2 T1PJR0 A0A182F1A4 A0A212EMX8 A0A182RWA4 A0A182Y9P5 A0A034W898 U4UJN8 A0A1A9WL19 A0A1I8NPE7 N6ULG0 J3JUC0 B0WNB3 A0A1A9XT88 A0A2S2NBB4 A0A0L7RI89 A0A182WRV7 A0A1A9ZCY6 A0A1Q3F1E5 C4WU15 A0A023EPR8 A0A232F1P4 A0A1B0FG51 A0A0A9YJR7 A0A1B6EBX6 A0A1B6L3U3 A0A1B6IV31 A0A2H8TMP7 T1HFB4 U5EIA3 A0A1L8DYQ4 A0A1J1HF19 A0A2S2QSF1 A0A2J7QJW4 A0A2A3E1T5 A0A0A1XRZ1 A0A0Q9XD10 B4KJQ9 A0A0M4E2G4 A0A3L8DPZ3 A0A026VVA5 A0A088AAE3 A0A195CJW5 A0A151X1F4 A0A195ETR6 A0A195DVJ3 F4W3Z5 B4LV86 E2APP5 B4JQW7 T1DPK7 A0A0J7NWS7 E2BBS8 A0A1A9UVE8 B4I563 A0A1A9WL14 B4N915 A0A336KUV6 A0A1B0AUP3 A0A1W4VXF8 A0A1W4VWK6
A0A1L8DYP3 A0A1B0GKX7 A0A1Y1M4L3 A0A2A4JG60 D6WN07 A0A1Y1M9I6 A0A0L0BS07 A0A1A9WL20 A0A2H1W2C5 W8BCS0 W8BHQ7 A0A0A1WFH5 A0A0A1WT83 A0A2C9GS57 A0A182MP58 A0A023F3V8 K7J1P8 A0A224XKX2 A0A1S4GYN2 A0A182UUY6 A0A182KMI3 A0A182UG21 A0A182IQG5 A0A0V0G9A4 A0A0K8VLN1 A0A182NTA5 A0A084VUV2 A0A182QW36 A0A0P6JSU2 A0A1I8ML52 A0A023ENN4 W5J4S0 A0A182P4K2 A0A0K8UZP2 T1PJR0 A0A182F1A4 A0A212EMX8 A0A182RWA4 A0A182Y9P5 A0A034W898 U4UJN8 A0A1A9WL19 A0A1I8NPE7 N6ULG0 J3JUC0 B0WNB3 A0A1A9XT88 A0A2S2NBB4 A0A0L7RI89 A0A182WRV7 A0A1A9ZCY6 A0A1Q3F1E5 C4WU15 A0A023EPR8 A0A232F1P4 A0A1B0FG51 A0A0A9YJR7 A0A1B6EBX6 A0A1B6L3U3 A0A1B6IV31 A0A2H8TMP7 T1HFB4 U5EIA3 A0A1L8DYQ4 A0A1J1HF19 A0A2S2QSF1 A0A2J7QJW4 A0A2A3E1T5 A0A0A1XRZ1 A0A0Q9XD10 B4KJQ9 A0A0M4E2G4 A0A3L8DPZ3 A0A026VVA5 A0A088AAE3 A0A195CJW5 A0A151X1F4 A0A195ETR6 A0A195DVJ3 F4W3Z5 B4LV86 E2APP5 B4JQW7 T1DPK7 A0A0J7NWS7 E2BBS8 A0A1A9UVE8 B4I563 A0A1A9WL14 B4N915 A0A336KUV6 A0A1B0AUP3 A0A1W4VXF8 A0A1W4VWK6
PDB
4KMS
E-value=4.62941e-19,
Score=231
Ontologies
GO
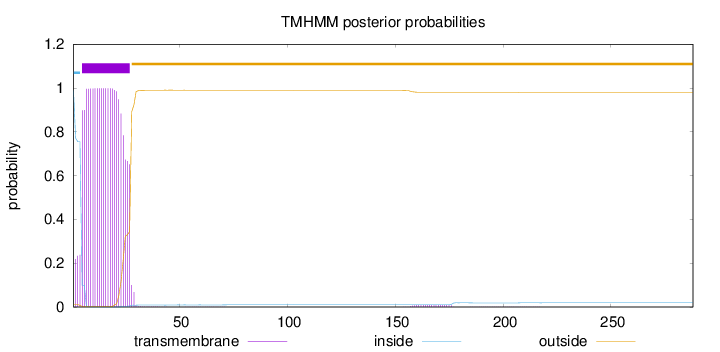
Topology
Length:
288
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.44306
Exp number, first 60 AAs:
22.24454
Total prob of N-in:
0.98912
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 288
Population Genetic Test Statistics
Pi
204.528702
Theta
176.582571
Tajima's D
0.485524
CLR
305.937241
CSRT
0.503574821258937
Interpretation
Uncertain
