Gene
KWMTBOMO01643
Pre Gene Modal
BGIBMGA006058
Annotation
PREDICTED:_sodium/potassium-transporting_ATPase_subunit_beta-1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.907 Nuclear Reliability : 4.656
Sequence
CDS
ATGCCTGAGAACGATACGACGGAGGAAAAGGAGTGGGTGAGAGGACCAGAGCACCTAGGCTCAGTATGGTCGAGGATAGCTCGAGCTATTTACAATCCTAAAGAAAGGAGCTTCTTGGGACGAACAGCCAAACGATGGGGTATCGTACTAGTATTCTACTTGGTATTTTACGCGGCGCTGGCGCTTCTATTCGGCATCTGCATGGCGGGTCTGGTGTACCACATTGATGCTACCACACCAACATACACGCTTGACCGATCCTTAATAGGTTCGAATCCAGGAGTGACCTACAGACCTAGACCAAATGACGGCATCAAAATCAGCATCGACATAAACAATTTCACTGAAACAAATGCGATTTTAAAAGATCTCGACGAGTTTTTTCAGCCTTACTTAATGGATAGCAGTTACAAATCGGAAAATGGTTGCAAATCTGACGACAATTACGGGTATCCCGACACGCCGTGCCTTTTCATAAAAATCAATAAGATATATGGTTGGACACCGGTTTATTACGAGAGCACAGATTTGCCAGCAGACATGCCGGAAGACTTAGTGGAGCATATCAAAGGATTAGATGCGAACGAGACTAAACGAGTATGGATATCGTGTTTAGAAGAGAAAGATAATGGCACTACCATAGAATATCCTTGGGGTCAAGGAATCCAGGGGAGCTCCTTCCCATTTTTGAACAAGGAAGGGCAGACGAGTCCTGTTGTAGCAGTGAGGATCACTCCCCCAGTGGGAAAGCTGATCGCGTTAAGATGCCGCGCGTGGGCAAAGAATATCCAATACCGCTTTAGCATCAAGGACTTCTCCGGATACACACCCATCCAGGTGGAAGTGGAAAACTCAGCCAACGTGACCGTCTAA
Protein
MPENDTTEEKEWVRGPEHLGSVWSRIARAIYNPKERSFLGRTAKRWGIVLVFYLVFYAALALLFGICMAGLVYHIDATTPTYTLDRSLIGSNPGVTYRPRPNDGIKISIDINNFTETNAILKDLDEFFQPYLMDSSYKSENGCKSDDNYGYPDTPCLFIKINKIYGWTPVYYESTDLPADMPEDLVEHIKGLDANETKRVWISCLEEKDNGTTIEYPWGQGIQGSSFPFLNKEGQTSPVVAVRITPPVGKLIALRCRAWAKNIQYRFSIKDFSGYTPIQVEVENSANVTV
Summary
Uniprot
H9J966
A0A2W1BI92
A0A194PYS8
A0A2H1V4D7
A0A212F3C3
A0A194R6I1
+ More
A0A182MCE8 K7IVU2 A0A232ETE4 T1PJY0 A0A084VI43 A0A1I8P7X3 W5JVD3 A0A1W4WPD8 U5ESK8 A0A182XZM9 A0A2H1VA10 A0A158P3Z0 A0A182J4Z7 B4KKY7 A0A0L0BL11 A0A195CNB9 A0A182PJP0 D3TLK7 B3MKP8 A0A182IEG1 A0A182HDG3 A0A182G6Y7 A0A3B0KIV7 A0A1Q3FKF7 A0A1W4UNI1 B0WIC2 A0A182Q2J7 A0A2M3Z8D9 W5U4X6 A0A1B0GFL3 A0A182TLK8 A0A182L2J9 A0A1S4GW77 A0A2M3Z881 A0A182VI54 B0WIC3 A0A182W7Q2 A0A026WRQ4 A0A2H8TWY8 A0A182RU89 A0NFA5 E2C796 A0A0V0G6C0 A0A2S2R3Y8 A0A023ENE1 E2AJ47 B4N1F5 A0A182JUG7 Q29P11 A0A194Q0F6 J9K155 A0A2H1VT27 A0A2W1BP20 B4LRJ8 A0A034V8P7 A0A194R6X6 A0A1Q3FKY4 A0A069DSA5 B4GJW9 A0A1B6IPD8 A0A1A9W5H5 A0A1B6C7U0 A0A1B6KK47 Q16TS1 A0A139WHY1 Q16TS0 A0A067R044 A0A088A4B1 A0A1B6FQY1 A0A151JVH5 A0A182MZB3 A0A0K8TYF7 A0A154NY21 A0A224XST4 A0A1B0CDT7 A0A212F3C2 A0A182X1I0 A0A0P4VNF2 A0A310SEI5 A0A1J1IMW2 A0A0A1WQG6 A8C9X0 D6WKU1 A0A0K8TRE2 A0A1A9ZFE5 A0A023F2Q2 W8AUT5 E0W230 E9J1Q8 A0A1A9VF44 A0A336M0F7 B4GU49 H9J968
A0A182MCE8 K7IVU2 A0A232ETE4 T1PJY0 A0A084VI43 A0A1I8P7X3 W5JVD3 A0A1W4WPD8 U5ESK8 A0A182XZM9 A0A2H1VA10 A0A158P3Z0 A0A182J4Z7 B4KKY7 A0A0L0BL11 A0A195CNB9 A0A182PJP0 D3TLK7 B3MKP8 A0A182IEG1 A0A182HDG3 A0A182G6Y7 A0A3B0KIV7 A0A1Q3FKF7 A0A1W4UNI1 B0WIC2 A0A182Q2J7 A0A2M3Z8D9 W5U4X6 A0A1B0GFL3 A0A182TLK8 A0A182L2J9 A0A1S4GW77 A0A2M3Z881 A0A182VI54 B0WIC3 A0A182W7Q2 A0A026WRQ4 A0A2H8TWY8 A0A182RU89 A0NFA5 E2C796 A0A0V0G6C0 A0A2S2R3Y8 A0A023ENE1 E2AJ47 B4N1F5 A0A182JUG7 Q29P11 A0A194Q0F6 J9K155 A0A2H1VT27 A0A2W1BP20 B4LRJ8 A0A034V8P7 A0A194R6X6 A0A1Q3FKY4 A0A069DSA5 B4GJW9 A0A1B6IPD8 A0A1A9W5H5 A0A1B6C7U0 A0A1B6KK47 Q16TS1 A0A139WHY1 Q16TS0 A0A067R044 A0A088A4B1 A0A1B6FQY1 A0A151JVH5 A0A182MZB3 A0A0K8TYF7 A0A154NY21 A0A224XST4 A0A1B0CDT7 A0A212F3C2 A0A182X1I0 A0A0P4VNF2 A0A310SEI5 A0A1J1IMW2 A0A0A1WQG6 A8C9X0 D6WKU1 A0A0K8TRE2 A0A1A9ZFE5 A0A023F2Q2 W8AUT5 E0W230 E9J1Q8 A0A1A9VF44 A0A336M0F7 B4GU49 H9J968
Pubmed
19121390
28756777
26354079
22118469
20075255
28648823
+ More
25315136 24438588 20920257 23761445 25244985 21347285 17994087 18057021 26108605 20353571 26483478 20966253 12364791 24508170 20798317 24945155 15632085 25348373 26334808 17510324 18362917 19820115 24845553 27129103 25830018 17760985 26369729 25474469 24495485 20566863 21282665
25315136 24438588 20920257 23761445 25244985 21347285 17994087 18057021 26108605 20353571 26483478 20966253 12364791 24508170 20798317 24945155 15632085 25348373 26334808 17510324 18362917 19820115 24845553 27129103 25830018 17760985 26369729 25474469 24495485 20566863 21282665
EMBL
BABH01004377
KZ150105
PZC73424.1
KQ459584
KPI98481.1
ODYU01000439
+ More
SOQ35212.1 AGBW02010580 OWR48237.1 KQ460644 KPJ13272.1 AXCM01003179 AXCM01003180 NNAY01002286 OXU21621.1 KA644528 KA649068 KA649071 AFP63700.1 ATLV01013278 ATLV01013279 KE524849 KFB37637.1 ADMH02000383 ETN66759.1 GANO01002307 JAB57564.1 ODYU01001458 SOQ37683.1 ADTU01008623 CH933807 EDW11717.1 KRG02859.1 KRG02860.1 KRG02861.1 JRES01001704 KNC20756.1 KQ977513 KYN02160.1 EZ422309 ADD18585.1 CH902620 EDV31579.1 APCN01005202 JXUM01034457 JXUM01034458 KQ561023 KXJ79991.1 JXUM01045990 JXUM01045991 KQ561458 KXJ78506.1 OUUW01000010 SPP85696.1 GFDL01006976 JAV28069.1 DS231946 EDS28367.1 AXCN02002070 AXCN02002071 GGFM01004021 MBW24772.1 KF813099 AHH35012.1 CCAG010007263 CCAG010007264 CCAG010007265 AAAB01008964 GGFM01003985 MBW24736.1 EDS28368.1 KK107119 EZA58730.1 GFXV01006506 MBW18311.1 EAU76401.1 GL453340 EFN76209.1 GECL01002590 JAP03534.1 GGMS01015451 MBY84654.1 JXUM01045994 JXUM01045995 JXUM01045996 GAPW01002671 JAC10927.1 KXJ78507.1 GL439967 EFN66487.1 CH963920 EDW78066.1 CH379058 EAL34482.1 KPI98479.1 ABLF02006785 ODYU01004281 SOQ43980.1 PZC73423.1 CH940649 EDW63593.1 GAKP01021024 JAC37928.1 KPJ13274.1 GFDL01006909 JAV28136.1 GBGD01002278 JAC86611.1 CH479184 EDW36935.1 GECU01018949 JAS88757.1 GEDC01027767 JAS09531.1 GEBQ01028156 JAT11821.1 CH477641 EAT37922.1 KQ971342 KYB27583.1 EAT37923.1 KK852836 KDR15243.1 GECZ01017189 JAS52580.1 KQ981701 KYN37502.1 GDHF01033026 JAI19288.1 KQ434782 KZC04575.1 GFTR01004834 JAW11592.1 AJWK01008216 OWR48238.1 GDKW01003308 JAI53287.1 KQ766043 OAD53763.1 CVRI01000054 CRL00884.1 GBXI01013431 GBXI01004631 JAD00861.1 JAD09661.1 EU032348 ABV44715.1 EFA02978.1 GDAI01000897 JAI16706.1 GBBI01003398 JAC15314.1 GAMC01013930 JAB92625.1 DS235874 EEB19686.1 GL767674 EFZ13236.1 UFQT01000377 SSX23725.1 CH479191 EDW26132.1
SOQ35212.1 AGBW02010580 OWR48237.1 KQ460644 KPJ13272.1 AXCM01003179 AXCM01003180 NNAY01002286 OXU21621.1 KA644528 KA649068 KA649071 AFP63700.1 ATLV01013278 ATLV01013279 KE524849 KFB37637.1 ADMH02000383 ETN66759.1 GANO01002307 JAB57564.1 ODYU01001458 SOQ37683.1 ADTU01008623 CH933807 EDW11717.1 KRG02859.1 KRG02860.1 KRG02861.1 JRES01001704 KNC20756.1 KQ977513 KYN02160.1 EZ422309 ADD18585.1 CH902620 EDV31579.1 APCN01005202 JXUM01034457 JXUM01034458 KQ561023 KXJ79991.1 JXUM01045990 JXUM01045991 KQ561458 KXJ78506.1 OUUW01000010 SPP85696.1 GFDL01006976 JAV28069.1 DS231946 EDS28367.1 AXCN02002070 AXCN02002071 GGFM01004021 MBW24772.1 KF813099 AHH35012.1 CCAG010007263 CCAG010007264 CCAG010007265 AAAB01008964 GGFM01003985 MBW24736.1 EDS28368.1 KK107119 EZA58730.1 GFXV01006506 MBW18311.1 EAU76401.1 GL453340 EFN76209.1 GECL01002590 JAP03534.1 GGMS01015451 MBY84654.1 JXUM01045994 JXUM01045995 JXUM01045996 GAPW01002671 JAC10927.1 KXJ78507.1 GL439967 EFN66487.1 CH963920 EDW78066.1 CH379058 EAL34482.1 KPI98479.1 ABLF02006785 ODYU01004281 SOQ43980.1 PZC73423.1 CH940649 EDW63593.1 GAKP01021024 JAC37928.1 KPJ13274.1 GFDL01006909 JAV28136.1 GBGD01002278 JAC86611.1 CH479184 EDW36935.1 GECU01018949 JAS88757.1 GEDC01027767 JAS09531.1 GEBQ01028156 JAT11821.1 CH477641 EAT37922.1 KQ971342 KYB27583.1 EAT37923.1 KK852836 KDR15243.1 GECZ01017189 JAS52580.1 KQ981701 KYN37502.1 GDHF01033026 JAI19288.1 KQ434782 KZC04575.1 GFTR01004834 JAW11592.1 AJWK01008216 OWR48238.1 GDKW01003308 JAI53287.1 KQ766043 OAD53763.1 CVRI01000054 CRL00884.1 GBXI01013431 GBXI01004631 JAD00861.1 JAD09661.1 EU032348 ABV44715.1 EFA02978.1 GDAI01000897 JAI16706.1 GBBI01003398 JAC15314.1 GAMC01013930 JAB92625.1 DS235874 EEB19686.1 GL767674 EFZ13236.1 UFQT01000377 SSX23725.1 CH479191 EDW26132.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000075883
UP000002358
+ More
UP000215335 UP000095301 UP000030765 UP000095300 UP000000673 UP000192223 UP000076408 UP000005205 UP000075880 UP000009192 UP000037069 UP000078542 UP000075885 UP000007801 UP000075840 UP000069940 UP000249989 UP000268350 UP000192221 UP000002320 UP000075886 UP000092444 UP000075902 UP000075882 UP000075903 UP000075920 UP000053097 UP000075900 UP000007062 UP000008237 UP000000311 UP000007798 UP000075881 UP000001819 UP000007819 UP000008792 UP000008744 UP000091820 UP000008820 UP000007266 UP000027135 UP000005203 UP000078541 UP000075884 UP000076502 UP000092461 UP000076407 UP000183832 UP000092445 UP000009046 UP000078200
UP000215335 UP000095301 UP000030765 UP000095300 UP000000673 UP000192223 UP000076408 UP000005205 UP000075880 UP000009192 UP000037069 UP000078542 UP000075885 UP000007801 UP000075840 UP000069940 UP000249989 UP000268350 UP000192221 UP000002320 UP000075886 UP000092444 UP000075902 UP000075882 UP000075903 UP000075920 UP000053097 UP000075900 UP000007062 UP000008237 UP000000311 UP000007798 UP000075881 UP000001819 UP000007819 UP000008792 UP000008744 UP000091820 UP000008820 UP000007266 UP000027135 UP000005203 UP000078541 UP000075884 UP000076502 UP000092461 UP000076407 UP000183832 UP000092445 UP000009046 UP000078200
Pfam
PF00287 Na_K-ATPase
Gene 3D
ProteinModelPortal
H9J966
A0A2W1BI92
A0A194PYS8
A0A2H1V4D7
A0A212F3C3
A0A194R6I1
+ More
A0A182MCE8 K7IVU2 A0A232ETE4 T1PJY0 A0A084VI43 A0A1I8P7X3 W5JVD3 A0A1W4WPD8 U5ESK8 A0A182XZM9 A0A2H1VA10 A0A158P3Z0 A0A182J4Z7 B4KKY7 A0A0L0BL11 A0A195CNB9 A0A182PJP0 D3TLK7 B3MKP8 A0A182IEG1 A0A182HDG3 A0A182G6Y7 A0A3B0KIV7 A0A1Q3FKF7 A0A1W4UNI1 B0WIC2 A0A182Q2J7 A0A2M3Z8D9 W5U4X6 A0A1B0GFL3 A0A182TLK8 A0A182L2J9 A0A1S4GW77 A0A2M3Z881 A0A182VI54 B0WIC3 A0A182W7Q2 A0A026WRQ4 A0A2H8TWY8 A0A182RU89 A0NFA5 E2C796 A0A0V0G6C0 A0A2S2R3Y8 A0A023ENE1 E2AJ47 B4N1F5 A0A182JUG7 Q29P11 A0A194Q0F6 J9K155 A0A2H1VT27 A0A2W1BP20 B4LRJ8 A0A034V8P7 A0A194R6X6 A0A1Q3FKY4 A0A069DSA5 B4GJW9 A0A1B6IPD8 A0A1A9W5H5 A0A1B6C7U0 A0A1B6KK47 Q16TS1 A0A139WHY1 Q16TS0 A0A067R044 A0A088A4B1 A0A1B6FQY1 A0A151JVH5 A0A182MZB3 A0A0K8TYF7 A0A154NY21 A0A224XST4 A0A1B0CDT7 A0A212F3C2 A0A182X1I0 A0A0P4VNF2 A0A310SEI5 A0A1J1IMW2 A0A0A1WQG6 A8C9X0 D6WKU1 A0A0K8TRE2 A0A1A9ZFE5 A0A023F2Q2 W8AUT5 E0W230 E9J1Q8 A0A1A9VF44 A0A336M0F7 B4GU49 H9J968
A0A182MCE8 K7IVU2 A0A232ETE4 T1PJY0 A0A084VI43 A0A1I8P7X3 W5JVD3 A0A1W4WPD8 U5ESK8 A0A182XZM9 A0A2H1VA10 A0A158P3Z0 A0A182J4Z7 B4KKY7 A0A0L0BL11 A0A195CNB9 A0A182PJP0 D3TLK7 B3MKP8 A0A182IEG1 A0A182HDG3 A0A182G6Y7 A0A3B0KIV7 A0A1Q3FKF7 A0A1W4UNI1 B0WIC2 A0A182Q2J7 A0A2M3Z8D9 W5U4X6 A0A1B0GFL3 A0A182TLK8 A0A182L2J9 A0A1S4GW77 A0A2M3Z881 A0A182VI54 B0WIC3 A0A182W7Q2 A0A026WRQ4 A0A2H8TWY8 A0A182RU89 A0NFA5 E2C796 A0A0V0G6C0 A0A2S2R3Y8 A0A023ENE1 E2AJ47 B4N1F5 A0A182JUG7 Q29P11 A0A194Q0F6 J9K155 A0A2H1VT27 A0A2W1BP20 B4LRJ8 A0A034V8P7 A0A194R6X6 A0A1Q3FKY4 A0A069DSA5 B4GJW9 A0A1B6IPD8 A0A1A9W5H5 A0A1B6C7U0 A0A1B6KK47 Q16TS1 A0A139WHY1 Q16TS0 A0A067R044 A0A088A4B1 A0A1B6FQY1 A0A151JVH5 A0A182MZB3 A0A0K8TYF7 A0A154NY21 A0A224XST4 A0A1B0CDT7 A0A212F3C2 A0A182X1I0 A0A0P4VNF2 A0A310SEI5 A0A1J1IMW2 A0A0A1WQG6 A8C9X0 D6WKU1 A0A0K8TRE2 A0A1A9ZFE5 A0A023F2Q2 W8AUT5 E0W230 E9J1Q8 A0A1A9VF44 A0A336M0F7 B4GU49 H9J968
PDB
5Y0B
E-value=0.000275596,
Score=103
Ontologies
GO
PANTHER
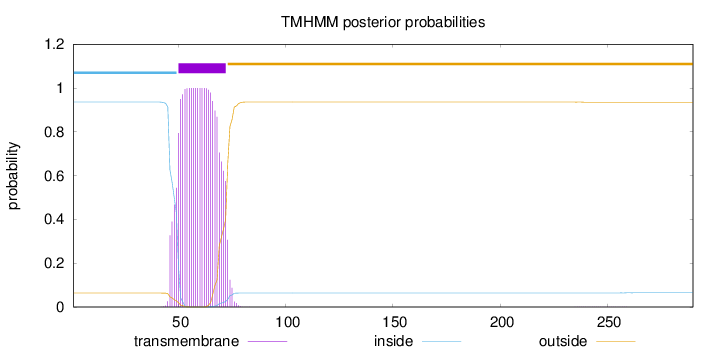
Topology
Length:
290
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.30913
Exp number, first 60 AAs:
12.47389
Total prob of N-in:
0.93622
POSSIBLE N-term signal
sequence
inside
1 - 49
TMhelix
50 - 72
outside
73 - 290
Population Genetic Test Statistics
Pi
266.38866
Theta
170.607924
Tajima's D
1.190384
CLR
0.34031
CSRT
0.709614519274036
Interpretation
Uncertain
