Gene
KWMTBOMO01641
Pre Gene Modal
BGIBMGA006099
Annotation
Na?K-ATPase_beta-subunit_[Bombyx_mori]
Full name
Sodium/potassium-transporting ATPase subunit beta-2
Alternative Name
Protein nervana 2
Sodium/potassium-dependent ATPase subunit beta-2
Sodium/potassium-dependent ATPase subunit beta-2
Location in the cell
Nuclear Reliability : 2.842
Sequence
CDS
ATGACGGTTAAGAAACGAACATCCGACCTGACGAGCTTAGAGCGATTCAACATGTACTACAGGGAGAAGGAACCACCTCTAACCAGATCTCAAAAAGTGAAACGGTTCATCTGGAACCCGAAGACCAGGCAGTTCTGTGGAAGAACAGGATCTAGTTGGTCAAAAATAGCATTGTTCTATTTTATCTTCTACTCCGCACTCGCCATCCTAGTCGCTATCTGCATGTGGACGTTCCTCCAGCTTTTGGATGCTAGGCAACCGAAATGGCAGCTCGAGAGGAGCATCATTGGCACTAACCCAGGGCTGGGCTTCAGGCCAACGCCTCCGGAGGTCGCCAGCAGTGTCATCTGGTACAAGGGAAATGACCCGGGTAGCCAACAGTTCTGGGTTAAAAAGCTATCCACCTTCTTAGCTGCGTACAAGCGCGATGGCAAAAAAGCTGGTGCGGGCCAGAACATCCACAACTGTGACTTCAAGCTTCCTCCTCCGGCTGGGAAAGTCTGCGATGTAGACATCAGCGCGTGGGGACCTTGCGTGCAGGACAACTATTTCGGCTACCACAAATCGACACCCTGCATCTTCCTGAAGCTAAATAAGATCTACGGTTGGAGGCCCAAGTTCTACAACAGCTCTGACAATCTACCCAAAGACATGCCGGAAGACTTGAAGGAGCACATCAGGAACATGACGGCGTATGATAAGAACTACCTAAACATGGTTTGGGTTTCCTGCCAAGGCGAGAACCCAGCTGATCGCGAGAACATTGGTCCCATACAGTACCTACCTCACCGAGGCTTCCCGGGATACTACTTCCCCTACACCAACCAAGAAGGATACCTGAGCCCTCTTGTAGCTGTGCACTTGCAGAGACCTAAGACTGGCATGTTAATCAACATTGAATGTCGTGCCTGGGCGAACAACATCAAGTATGAGAGACTTGAAGCCATGGGCTCTGTACACATCGAGATGTTGATCGAGTAG
Protein
MTVKKRTSDLTSLERFNMYYREKEPPLTRSQKVKRFIWNPKTRQFCGRTGSSWSKIALFYFIFYSALAILVAICMWTFLQLLDARQPKWQLERSIIGTNPGLGFRPTPPEVASSVIWYKGNDPGSQQFWVKKLSTFLAAYKRDGKKAGAGQNIHNCDFKLPPPAGKVCDVDISAWGPCVQDNYFGYHKSTPCIFLKLNKIYGWRPKFYNSSDNLPKDMPEDLKEHIRNMTAYDKNYLNMVWVSCQGENPADRENIGPIQYLPHRGFPGYYFPYTNQEGYLSPLVAVHLQRPKTGMLINIECRAWANNIKYERLEAMGSVHIEMLIE
Summary
Description
This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The beta subunit regulates, through assembly of alpha/beta heterodimers, the number of sodium pumps transported to the plasma membrane.
Subunit
The sodium/potassium-transporting ATPase is composed of a catalytic alpha subunit, an auxiliary non-catalytic beta subunit and an additional regulatory subunit.
Similarity
Belongs to the X(+)/potassium ATPases subunit beta family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Ion transport
Membrane
Potassium
Potassium transport
Reference proteome
Signal-anchor
Sodium
Sodium transport
Transmembrane
Transmembrane helix
Transport
Feature
chain Sodium/potassium-transporting ATPase subunit beta-2
splice variant In isoform 2.1.
splice variant In isoform 2.1.
Uniprot
A0A0K2S4T7
A0A194Q0F6
A0A2W1BE21
A0A2A4K0R6
A0A2H1VT27
A0A212F3C0
+ More
A0A194R6X6 H9J9A7 A0A3L8DD16 A0A151JW23 E9J1Q7 A0A151X0L1 A0A195BJK0 A0A158P3Y8 E2C795 F4WKT5 E2AJ48 A0A026WUE4 A0A195CN39 A0A151JM32 A0A336M5T7 A0A194PYR8 A0A2H1V2Z3 V9IJT5 A0A088A4D5 A0A310S7A7 A0A034VPI2 A0A154NY27 W8AQQ7 A0A0A1XIS6 A0A2C9H707 Q7Q3B0 A0A0L7QPX7 B4LRJ7 Q16TR9 T1D4D1 A0A1L8DFI6 A0A1Q3G4C7 B4JB94 A0A0L0BL02 B4KKY6 T1PK16 A0A232ETF2 K7IVU1 A0A182URH5 A0A182X1I1 A0A2W1BI97 A0A0K8W790 B4N1F6 A0A182L2K0 A0A1S4GW98 A0A1I8Q9N6 A0A034VNN3 E0VS44 W5U5U7 A0A182QHA3 A0A0Q9WFY6 A0A2A4JHJ4 U5EXA6 A0A212F3C8 A0A182JE55 A0A336MCH0 B4P009 A0A0A1XLF2 A4V0B5 Q24048 J9EAN8 W8ALM6 A0A0Q9X198 A0A0J9THC4 B4HXY0 A0A023EQW5 A0A1S4JIQ4 B3N5X3 B3MKP9 A0A2M4CPQ7 A0A1B0FFL4 A0A1W4UQD7 A0A2M3Z1Y5 A0A0Q9XBM1 A0A2M4CPK3 A0A1L8DF34 T1EB20 A0A2M4AFG7 A0A182HDG2 Q24048-2 Q1RL12 A0A182FNA7 A0A0J9QX06 Q29P10 B4GJW8 A0A182XZM8 A0A0R1DR04 A0A067RA63 A0A2M4BU32 A0A1Q3FIL8 A0A1W4UNJ0 A0A0Q5WAH7 A0A0N8NZ69 A0A3B0JUB9
A0A194R6X6 H9J9A7 A0A3L8DD16 A0A151JW23 E9J1Q7 A0A151X0L1 A0A195BJK0 A0A158P3Y8 E2C795 F4WKT5 E2AJ48 A0A026WUE4 A0A195CN39 A0A151JM32 A0A336M5T7 A0A194PYR8 A0A2H1V2Z3 V9IJT5 A0A088A4D5 A0A310S7A7 A0A034VPI2 A0A154NY27 W8AQQ7 A0A0A1XIS6 A0A2C9H707 Q7Q3B0 A0A0L7QPX7 B4LRJ7 Q16TR9 T1D4D1 A0A1L8DFI6 A0A1Q3G4C7 B4JB94 A0A0L0BL02 B4KKY6 T1PK16 A0A232ETF2 K7IVU1 A0A182URH5 A0A182X1I1 A0A2W1BI97 A0A0K8W790 B4N1F6 A0A182L2K0 A0A1S4GW98 A0A1I8Q9N6 A0A034VNN3 E0VS44 W5U5U7 A0A182QHA3 A0A0Q9WFY6 A0A2A4JHJ4 U5EXA6 A0A212F3C8 A0A182JE55 A0A336MCH0 B4P009 A0A0A1XLF2 A4V0B5 Q24048 J9EAN8 W8ALM6 A0A0Q9X198 A0A0J9THC4 B4HXY0 A0A023EQW5 A0A1S4JIQ4 B3N5X3 B3MKP9 A0A2M4CPQ7 A0A1B0FFL4 A0A1W4UQD7 A0A2M3Z1Y5 A0A0Q9XBM1 A0A2M4CPK3 A0A1L8DF34 T1EB20 A0A2M4AFG7 A0A182HDG2 Q24048-2 Q1RL12 A0A182FNA7 A0A0J9QX06 Q29P10 B4GJW8 A0A182XZM8 A0A0R1DR04 A0A067RA63 A0A2M4BU32 A0A1Q3FIL8 A0A1W4UNJ0 A0A0Q5WAH7 A0A0N8NZ69 A0A3B0JUB9
Pubmed
26354079
28756777
22118469
19121390
30249741
21282665
+ More
21347285 20798317 21719571 24508170 25348373 24495485 25830018 12364791 14747013 17210077 17994087 17510324 24330624 26108605 25315136 28648823 20075255 20966253 20566863 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7777518 12537569 7540667 9648860 17893096 22936249 24945155 18057021 26483478 15632085 25244985 24845553
21347285 20798317 21719571 24508170 25348373 24495485 25830018 12364791 14747013 17210077 17994087 17510324 24330624 26108605 25315136 28648823 20075255 20966253 20566863 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7777518 12537569 7540667 9648860 17893096 22936249 24945155 18057021 26483478 15632085 25244985 24845553
EMBL
LC029031
BAS22118.1
KQ459584
KPI98479.1
KZ150105
PZC73422.1
+ More
NWSH01000323 PCG77484.1 ODYU01004281 SOQ43980.1 AGBW02010580 OWR48239.1 KQ460644 KPJ13274.1 BABH01004376 BABH01004377 QOIP01000009 RLU18375.1 KQ981701 KYN37501.1 GL767674 EFZ13198.1 KQ982612 KYQ53877.1 KQ976453 KYM85348.1 ADTU01008623 GL453340 EFN76208.1 GL888206 EGI65177.1 GL439967 EFN66488.1 KK107119 EZA58729.1 KQ977513 KYN02161.1 KQ978949 KYN27360.1 UFQT01000377 SSX23727.1 KPI98482.1 ODYU01000439 SOQ35210.1 JR049281 AEY60917.1 KQ766043 OAD53764.1 GAKP01015263 GAKP01015262 JAC43690.1 KQ434782 KZC04576.1 GAMC01019727 JAB86828.1 GBXI01005074 GBXI01003386 JAD09218.1 JAD10906.1 AAAB01008964 EAA12801.3 KQ414813 KOC60536.1 CH940649 EDW63592.1 CH477641 EAT37924.1 EAT37926.1 GALA01001003 JAA93849.1 GFDF01008939 JAV05145.1 GFDL01000429 JAV34616.1 CH916368 EDW02899.1 JRES01001704 KNC20746.1 CH933807 EDW11716.1 KA649142 AFP63771.1 NNAY01002286 OXU21622.1 PZC73425.1 GDHF01030989 GDHF01028963 GDHF01005372 JAI21325.1 JAI23351.1 JAI46942.1 CH963920 EDW78067.1 GAKP01015261 GAKP01015260 GAKP01015259 JAC43692.1 DS235744 EEB16200.1 KF813100 AHH35013.1 AXCN02002071 AXCN02002072 KRF81175.1 NWSH01001558 PCG70862.1 GANO01002628 JAB57243.1 OWR48236.1 SSX23728.1 CM000157 EDW87836.1 KRJ97426.1 KRJ97427.1 GBXI01006452 GBXI01004712 GBXI01002506 JAD07840.1 JAD09580.1 JAD11786.1 AE014134 AAX52659.1 AGB92700.1 U22439 U22440 AY060289 EJY57851.1 GAMC01019728 JAB86827.1 KRF98806.1 CM002910 KMY88625.1 CH480818 EDW51910.1 GAPW01002579 JAC11019.1 CH954177 EDV59132.1 CH902620 EDV31580.1 KPU73493.1 KPU73494.1 GGFL01003105 MBW67283.1 CCAG010002985 CCAG010002986 GGFM01001775 MBW22526.1 KRG02858.1 GGFL01003076 MBW67254.1 GFDF01009089 JAV04995.1 GAMD01000463 JAB01128.1 GGFK01006218 MBW39539.1 JXUM01034452 JXUM01034453 JXUM01034454 JXUM01034455 JXUM01034456 KQ561023 KXJ79990.1 BT025048 AAF52439.1 AAX52660.1 ABE73219.1 KMY88624.1 KMY88626.1 KMY88627.1 KMY88628.1 CH379058 EAL34483.1 CH479184 EDW36934.1 KRJ97425.1 KK852595 KDR20644.1 GGFJ01007372 MBW56513.1 GFDL01007628 JAV27417.1 KQS70647.1 KPU73492.1 OUUW01000010 SPP85694.1
NWSH01000323 PCG77484.1 ODYU01004281 SOQ43980.1 AGBW02010580 OWR48239.1 KQ460644 KPJ13274.1 BABH01004376 BABH01004377 QOIP01000009 RLU18375.1 KQ981701 KYN37501.1 GL767674 EFZ13198.1 KQ982612 KYQ53877.1 KQ976453 KYM85348.1 ADTU01008623 GL453340 EFN76208.1 GL888206 EGI65177.1 GL439967 EFN66488.1 KK107119 EZA58729.1 KQ977513 KYN02161.1 KQ978949 KYN27360.1 UFQT01000377 SSX23727.1 KPI98482.1 ODYU01000439 SOQ35210.1 JR049281 AEY60917.1 KQ766043 OAD53764.1 GAKP01015263 GAKP01015262 JAC43690.1 KQ434782 KZC04576.1 GAMC01019727 JAB86828.1 GBXI01005074 GBXI01003386 JAD09218.1 JAD10906.1 AAAB01008964 EAA12801.3 KQ414813 KOC60536.1 CH940649 EDW63592.1 CH477641 EAT37924.1 EAT37926.1 GALA01001003 JAA93849.1 GFDF01008939 JAV05145.1 GFDL01000429 JAV34616.1 CH916368 EDW02899.1 JRES01001704 KNC20746.1 CH933807 EDW11716.1 KA649142 AFP63771.1 NNAY01002286 OXU21622.1 PZC73425.1 GDHF01030989 GDHF01028963 GDHF01005372 JAI21325.1 JAI23351.1 JAI46942.1 CH963920 EDW78067.1 GAKP01015261 GAKP01015260 GAKP01015259 JAC43692.1 DS235744 EEB16200.1 KF813100 AHH35013.1 AXCN02002071 AXCN02002072 KRF81175.1 NWSH01001558 PCG70862.1 GANO01002628 JAB57243.1 OWR48236.1 SSX23728.1 CM000157 EDW87836.1 KRJ97426.1 KRJ97427.1 GBXI01006452 GBXI01004712 GBXI01002506 JAD07840.1 JAD09580.1 JAD11786.1 AE014134 AAX52659.1 AGB92700.1 U22439 U22440 AY060289 EJY57851.1 GAMC01019728 JAB86827.1 KRF98806.1 CM002910 KMY88625.1 CH480818 EDW51910.1 GAPW01002579 JAC11019.1 CH954177 EDV59132.1 CH902620 EDV31580.1 KPU73493.1 KPU73494.1 GGFL01003105 MBW67283.1 CCAG010002985 CCAG010002986 GGFM01001775 MBW22526.1 KRG02858.1 GGFL01003076 MBW67254.1 GFDF01009089 JAV04995.1 GAMD01000463 JAB01128.1 GGFK01006218 MBW39539.1 JXUM01034452 JXUM01034453 JXUM01034454 JXUM01034455 JXUM01034456 KQ561023 KXJ79990.1 BT025048 AAF52439.1 AAX52660.1 ABE73219.1 KMY88624.1 KMY88626.1 KMY88627.1 KMY88628.1 CH379058 EAL34483.1 CH479184 EDW36934.1 KRJ97425.1 KK852595 KDR20644.1 GGFJ01007372 MBW56513.1 GFDL01007628 JAV27417.1 KQS70647.1 KPU73492.1 OUUW01000010 SPP85694.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000053240
UP000005204
UP000279307
+ More
UP000078541 UP000075809 UP000078540 UP000005205 UP000008237 UP000007755 UP000000311 UP000053097 UP000078542 UP000078492 UP000005203 UP000076502 UP000076407 UP000007062 UP000053825 UP000008792 UP000008820 UP000001070 UP000037069 UP000009192 UP000095301 UP000215335 UP000002358 UP000075903 UP000007798 UP000075882 UP000095300 UP000009046 UP000075886 UP000075880 UP000002282 UP000000803 UP000001292 UP000008711 UP000007801 UP000092444 UP000192221 UP000069940 UP000249989 UP000069272 UP000001819 UP000008744 UP000076408 UP000027135 UP000268350
UP000078541 UP000075809 UP000078540 UP000005205 UP000008237 UP000007755 UP000000311 UP000053097 UP000078542 UP000078492 UP000005203 UP000076502 UP000076407 UP000007062 UP000053825 UP000008792 UP000008820 UP000001070 UP000037069 UP000009192 UP000095301 UP000215335 UP000002358 UP000075903 UP000007798 UP000075882 UP000095300 UP000009046 UP000075886 UP000075880 UP000002282 UP000000803 UP000001292 UP000008711 UP000007801 UP000092444 UP000192221 UP000069940 UP000249989 UP000069272 UP000001819 UP000008744 UP000076408 UP000027135 UP000268350
Pfam
PF00287 Na_K-ATPase
Gene 3D
ProteinModelPortal
A0A0K2S4T7
A0A194Q0F6
A0A2W1BE21
A0A2A4K0R6
A0A2H1VT27
A0A212F3C0
+ More
A0A194R6X6 H9J9A7 A0A3L8DD16 A0A151JW23 E9J1Q7 A0A151X0L1 A0A195BJK0 A0A158P3Y8 E2C795 F4WKT5 E2AJ48 A0A026WUE4 A0A195CN39 A0A151JM32 A0A336M5T7 A0A194PYR8 A0A2H1V2Z3 V9IJT5 A0A088A4D5 A0A310S7A7 A0A034VPI2 A0A154NY27 W8AQQ7 A0A0A1XIS6 A0A2C9H707 Q7Q3B0 A0A0L7QPX7 B4LRJ7 Q16TR9 T1D4D1 A0A1L8DFI6 A0A1Q3G4C7 B4JB94 A0A0L0BL02 B4KKY6 T1PK16 A0A232ETF2 K7IVU1 A0A182URH5 A0A182X1I1 A0A2W1BI97 A0A0K8W790 B4N1F6 A0A182L2K0 A0A1S4GW98 A0A1I8Q9N6 A0A034VNN3 E0VS44 W5U5U7 A0A182QHA3 A0A0Q9WFY6 A0A2A4JHJ4 U5EXA6 A0A212F3C8 A0A182JE55 A0A336MCH0 B4P009 A0A0A1XLF2 A4V0B5 Q24048 J9EAN8 W8ALM6 A0A0Q9X198 A0A0J9THC4 B4HXY0 A0A023EQW5 A0A1S4JIQ4 B3N5X3 B3MKP9 A0A2M4CPQ7 A0A1B0FFL4 A0A1W4UQD7 A0A2M3Z1Y5 A0A0Q9XBM1 A0A2M4CPK3 A0A1L8DF34 T1EB20 A0A2M4AFG7 A0A182HDG2 Q24048-2 Q1RL12 A0A182FNA7 A0A0J9QX06 Q29P10 B4GJW8 A0A182XZM8 A0A0R1DR04 A0A067RA63 A0A2M4BU32 A0A1Q3FIL8 A0A1W4UNJ0 A0A0Q5WAH7 A0A0N8NZ69 A0A3B0JUB9
A0A194R6X6 H9J9A7 A0A3L8DD16 A0A151JW23 E9J1Q7 A0A151X0L1 A0A195BJK0 A0A158P3Y8 E2C795 F4WKT5 E2AJ48 A0A026WUE4 A0A195CN39 A0A151JM32 A0A336M5T7 A0A194PYR8 A0A2H1V2Z3 V9IJT5 A0A088A4D5 A0A310S7A7 A0A034VPI2 A0A154NY27 W8AQQ7 A0A0A1XIS6 A0A2C9H707 Q7Q3B0 A0A0L7QPX7 B4LRJ7 Q16TR9 T1D4D1 A0A1L8DFI6 A0A1Q3G4C7 B4JB94 A0A0L0BL02 B4KKY6 T1PK16 A0A232ETF2 K7IVU1 A0A182URH5 A0A182X1I1 A0A2W1BI97 A0A0K8W790 B4N1F6 A0A182L2K0 A0A1S4GW98 A0A1I8Q9N6 A0A034VNN3 E0VS44 W5U5U7 A0A182QHA3 A0A0Q9WFY6 A0A2A4JHJ4 U5EXA6 A0A212F3C8 A0A182JE55 A0A336MCH0 B4P009 A0A0A1XLF2 A4V0B5 Q24048 J9EAN8 W8ALM6 A0A0Q9X198 A0A0J9THC4 B4HXY0 A0A023EQW5 A0A1S4JIQ4 B3N5X3 B3MKP9 A0A2M4CPQ7 A0A1B0FFL4 A0A1W4UQD7 A0A2M3Z1Y5 A0A0Q9XBM1 A0A2M4CPK3 A0A1L8DF34 T1EB20 A0A2M4AFG7 A0A182HDG2 Q24048-2 Q1RL12 A0A182FNA7 A0A0J9QX06 Q29P10 B4GJW8 A0A182XZM8 A0A0R1DR04 A0A067RA63 A0A2M4BU32 A0A1Q3FIL8 A0A1W4UNJ0 A0A0Q5WAH7 A0A0N8NZ69 A0A3B0JUB9
PDB
4XE5
E-value=6.77949e-29,
Score=316
Ontologies
GO
GO:0005890
GO:0006814
GO:0006813
GO:0016021
GO:0001671
GO:0030007
GO:0006883
GO:1990573
GO:0010248
GO:0036376
GO:0035158
GO:0060857
GO:0007605
GO:0030424
GO:0061343
GO:0019991
GO:0005391
GO:0005918
GO:0035159
GO:0035151
GO:0005886
GO:0098655
GO:0006812
GO:0007424
GO:0004190
GO:0006508
GO:0006811
GO:0016020
GO:0005515
GO:0003779
PANTHER
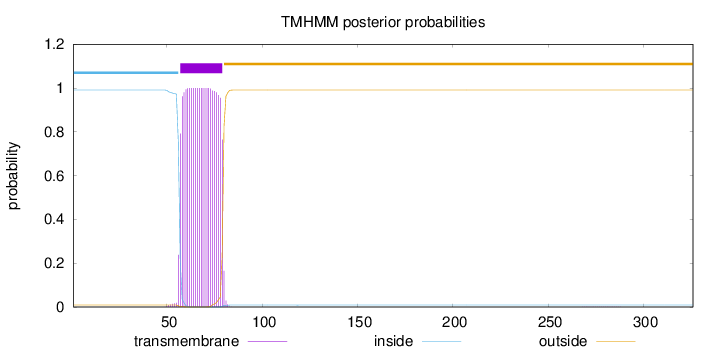
Topology
Subcellular location
Cell membrane
Length:
326
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.88913
Exp number, first 60 AAs:
4.03857
Total prob of N-in:
0.99121
inside
1 - 56
TMhelix
57 - 79
outside
80 - 326
Population Genetic Test Statistics
Pi
201.236599
Theta
163.043747
Tajima's D
0.706672
CLR
0
CSRT
0.576421178941053
Interpretation
Uncertain
