Pre Gene Modal
BGIBMGA006062
Annotation
PREDICTED:_sodium/potassium-transporting_ATPase_subunit_beta-2-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.069
Sequence
CDS
ATGGCGGACAAGAACAAAGTAGCTGAACAGTATTACGCTCCGCCACCGGACCTAGGGAAATGGGAGGCGTTCAGGATCTTCCTGTACAACTCGGAGACGGGCCAGGTCCTCGGGCGCACAGGCTCAAGCTGGGCAAAGATCCTGCTATTTTACCTTATCTTCTACGCGATTTTGGTGGGCTTCTTCGCTGCTCTGCTAGCGGTATTCTACCAGACCCTGGACACCAAGGTGCCGAAGTGGCAGCTTGATAGTTCAATAATCGGCAGTAATCCAGGTCTGGGCTTCAGGCCAATGCCAGATACTGCCAACGTGGAAAGTACTCTGATCTATTACAAGGTCAACGATAAGGGATCTGTGCTCAAATGGGCCAGGGTTATTGACGAATTTTTGAATCAATACCGTAAGAAAGGCAGCGGCAGTGGTGAAGCTCATGGTGCCGAAAACCGCGTACCATGCTCTCCCAGCAGCGGACCACTCGGCGAGAAGCAAGTCTGTGATGTACCAGTCGATGACTTCAATCCCTGCACTCCAGCCAACCAGTACAATTACGAACAAGCCGGCCCTTGCGTCTTCCTCAAGCTCAATAAGATATACAACTGGAGGCCGCAGCCGTACAACAACACCGAGGACCTACCCACGAACATGCCCGAGGATCTCAAGCAGCACATCAAGACACTGTCTGGTAAACCCGACGCCAACACTGTGTGGGTATCCTGTGAAGGCGAGAACCCTGCTGACGTGGAGAACATCGGCCCAGTCCAGTACATCCCCCGACGCGGGTTCCCTTCCTACTACTACCCGTTCACCAACAAGGAGGGATATCTAAGCCCACTCGTTGCTGTACTCTTTGAGAAACCTAGAACCGGGGTCCTCATCAACATCGAGTGCAAAGCCTGGGCGAAGAACATCTTCTACGACCGCTACGAGAGACGCGGCTCAGTCCACTTCGAGCTGATGGTGGACCGCGCTTAA
Protein
MADKNKVAEQYYAPPPDLGKWEAFRIFLYNSETGQVLGRTGSSWAKILLFYLIFYAILVGFFAALLAVFYQTLDTKVPKWQLDSSIIGSNPGLGFRPMPDTANVESTLIYYKVNDKGSVLKWARVIDEFLNQYRKKGSGSGEAHGAENRVPCSPSSGPLGEKQVCDVPVDDFNPCTPANQYNYEQAGPCVFLKLNKIYNWRPQPYNNTEDLPTNMPEDLKQHIKTLSGKPDANTVWVSCEGENPADVENIGPVQYIPRRGFPSYYYPFTNKEGYLSPLVAVLFEKPRTGVLINIECKAWAKNIFYDRYERRGSVHFELMVDRA
Summary
Uniprot
H9J970
A0A2H1VU14
A0A2W1BL15
A0A2A4K154
A0A194RBK0
A0A194PYR3
+ More
A0A212F3C9 A0A0L7LDP8 U5EVF5 K7IVU0 A0A0J7L1W9 E2AJ49 A0A088A4D4 A0A026WRS7 E2C794 A0A2A3E9E4 V9I8J8 A0A310SHK5 A0A232F5D8 A0A067RM12 A0A0C9RVD0 A0A0L7QPH5 A0A154PEL0 A0A195BKU6 A0A151X0X1 T1E275 F4WKT6 A0A151JVT6 A0A151JM06 A0A182IDW3 Q7PK91 A0A1Q3FYT7 A0A1Q3FYQ3 A0A023EPB7 T1E8B1 A0A182V944 Q16RY8 A0A182QDC3 E9J1Q5 A0A158P3Y7 A0A2M4A6M2 A0A2M3Z0F1 A0A2M4CTN4 W5UB64 A0A2M4BUW5 A0A336LVH5 A0A1B6KSW6 A0A182NUX5 A0A1L8DEZ6 E0VVM0 A0A195CN73 A0A1B6HRM5 A0A1L8DF72 A0A1B6J844 A0A182Y5L6 A0A1S4JYY7 A0A1D1Y1U2 A0A2J7RL52 A0A0A9Z1J2 A0A2J7RL67 A0A1Y1KPJ3 B4M9T2 A0A0P4VU23 R4FNJ2 U4U4L8 A0A0Q9VZG3 W5U5U7 A0A0M4EQG5 A0A0V0G820 B4P6W4 A0A023F8Y2 Q16TR9 Q86NM2 W8B1U8 B4JPW9 A0A1B6D385 A0A0J9R6J8 B4IFP0 B3NLW8 Q7JS69 A0A0R1DPA3 A0A1L8EHL9 D3TLK5 W8AQD4 A0A1W4UY15 A0A0J9TSR4 M9PDX8 A0A0Q5VXC3 B4KEZ8 A0A182JE55 A0A2C9H707 B4N7Q8 A0A0A1XBW7 Q7Q3B0 A0A0Q9X4L1 A0A0K8W9K6 E2AJ48 A0A034VN44 A0A0A1WGW0 B3MMB1
A0A212F3C9 A0A0L7LDP8 U5EVF5 K7IVU0 A0A0J7L1W9 E2AJ49 A0A088A4D4 A0A026WRS7 E2C794 A0A2A3E9E4 V9I8J8 A0A310SHK5 A0A232F5D8 A0A067RM12 A0A0C9RVD0 A0A0L7QPH5 A0A154PEL0 A0A195BKU6 A0A151X0X1 T1E275 F4WKT6 A0A151JVT6 A0A151JM06 A0A182IDW3 Q7PK91 A0A1Q3FYT7 A0A1Q3FYQ3 A0A023EPB7 T1E8B1 A0A182V944 Q16RY8 A0A182QDC3 E9J1Q5 A0A158P3Y7 A0A2M4A6M2 A0A2M3Z0F1 A0A2M4CTN4 W5UB64 A0A2M4BUW5 A0A336LVH5 A0A1B6KSW6 A0A182NUX5 A0A1L8DEZ6 E0VVM0 A0A195CN73 A0A1B6HRM5 A0A1L8DF72 A0A1B6J844 A0A182Y5L6 A0A1S4JYY7 A0A1D1Y1U2 A0A2J7RL52 A0A0A9Z1J2 A0A2J7RL67 A0A1Y1KPJ3 B4M9T2 A0A0P4VU23 R4FNJ2 U4U4L8 A0A0Q9VZG3 W5U5U7 A0A0M4EQG5 A0A0V0G820 B4P6W4 A0A023F8Y2 Q16TR9 Q86NM2 W8B1U8 B4JPW9 A0A1B6D385 A0A0J9R6J8 B4IFP0 B3NLW8 Q7JS69 A0A0R1DPA3 A0A1L8EHL9 D3TLK5 W8AQD4 A0A1W4UY15 A0A0J9TSR4 M9PDX8 A0A0Q5VXC3 B4KEZ8 A0A182JE55 A0A2C9H707 B4N7Q8 A0A0A1XBW7 Q7Q3B0 A0A0Q9X4L1 A0A0K8W9K6 E2AJ48 A0A034VN44 A0A0A1WGW0 B3MMB1
Pubmed
19121390
28756777
26354079
22118469
26227816
20075255
+ More
20798317 24508170 30249741 28648823 24845553 24330624 21719571 12364791 14747013 17210077 24945155 17510324 21282665 21347285 20566863 25244985 25401762 26823975 28004739 17994087 27129103 23537049 18057021 17550304 25474469 24495485 22936249 10731132 12537568 12537572 12537573 12537574 12930776 16110336 17569856 17569867 26109357 26109356 20353571 25830018 25348373
20798317 24508170 30249741 28648823 24845553 24330624 21719571 12364791 14747013 17210077 24945155 17510324 21282665 21347285 20566863 25244985 25401762 26823975 28004739 17994087 27129103 23537049 18057021 17550304 25474469 24495485 22936249 10731132 12537568 12537572 12537573 12537574 12930776 16110336 17569856 17569867 26109357 26109356 20353571 25830018 25348373
EMBL
BABH01004374
BABH01004375
ODYU01004281
SOQ43982.1
KZ150105
PZC73420.1
+ More
NWSH01000323 PCG77483.1 KQ460644 KPJ13276.1 KQ459584 KPI98477.1 AGBW02010580 OWR48241.1 JTDY01001561 KOB73524.1 GANO01001901 JAB57970.1 LBMM01001166 KMQ96792.1 GL439967 EFN66489.1 KK107119 QOIP01000009 EZA58727.1 RLU18942.1 GL453340 EFN76207.1 KZ288344 PBC27681.1 JR035723 JR035724 AEY56633.1 AEY56634.1 KQ766043 OAD53765.1 NNAY01000939 OXU25822.1 KK852595 KDR20642.1 GBYB01012845 JAG82612.1 KQ414813 KOC60537.1 KQ434874 KZC09650.1 KQ976453 KYM85345.1 KQ982612 KYQ53875.1 GALA01000989 JAA93863.1 GL888206 EGI65178.1 KQ981701 KYN37500.1 KQ978949 KYN27359.1 APCN01004534 APCN01004535 AAAB01008980 EAA43463.4 GFDL01002339 JAV32706.1 GFDL01002340 JAV32705.1 GAPW01002763 JAC10835.1 GAMD01002332 JAA99258.1 CH477692 EAT37221.1 AXCN02000751 AXCN02000752 GL767674 EFZ13266.1 ADTU01008618 ADTU01008619 ADTU01008620 GGFK01002957 MBW36278.1 GGFM01001261 MBW22012.1 GGFL01004447 MBW68625.1 KF813098 AHH35011.1 GGFJ01007600 MBW56741.1 UFQT01000215 SSX21830.1 GEBQ01025457 JAT14520.1 GFDF01009051 JAV05033.1 DS235812 EEB17426.1 KQ977513 KYN02163.1 GECU01030411 JAS77295.1 GFDF01009049 JAV05035.1 GECU01012359 JAS95347.1 GDJX01019345 JAT48591.1 NEVH01002699 PNF41568.1 GBHO01005255 GBRD01000598 GDHC01008674 JAG38349.1 JAG65223.1 JAQ09955.1 PNF41573.1 GEZM01077309 JAV63332.1 CH940654 EDW57958.1 GDKW01001068 JAI55527.1 GAHY01000563 JAA76947.1 KB631641 ERL84900.1 KRF78050.1 KRF78051.1 KF813100 AHH35013.1 CP012523 ALC39250.1 GECL01001960 JAP04164.1 CM000158 EDW89933.1 GBBI01000815 JAC17897.1 CH477641 EAT37924.1 EAT37926.1 BT004839 AAO45195.1 GAMC01015562 GAMC01015561 JAB90993.1 CH916372 EDV98949.1 GEDC01017263 JAS20035.1 CM002910 KMY91349.1 CH480833 EDW46462.1 CH954179 EDV54434.1 AF202633 AE014134 AY314744 BT044501 AAF17587.1 AAF53995.1 AAN11121.1 AAN11122.1 AAP79432.1 ACH95275.1 KRJ98796.1 KRJ98797.1 KRJ98798.1 GFDG01000654 JAV18145.1 EZ422307 ADD18583.1 GAMC01015560 JAB90995.1 KMY91350.1 AGB93210.1 KQS61786.1 CH933807 EDW11967.1 CH964214 EDW80397.1 GBXI01005852 JAD08440.1 AAAB01008964 EAA12801.3 KRG03005.1 KRG03006.1 KRG03007.1 GDHF01015849 GDHF01014106 GDHF01013089 GDHF01004473 GDHF01001608 JAI36465.1 JAI38208.1 JAI39225.1 JAI47841.1 JAI50706.1 EFN66488.1 GAKP01015964 GAKP01015962 JAC42988.1 GBXI01016170 JAC98121.1 CH902620 EDV31871.1
NWSH01000323 PCG77483.1 KQ460644 KPJ13276.1 KQ459584 KPI98477.1 AGBW02010580 OWR48241.1 JTDY01001561 KOB73524.1 GANO01001901 JAB57970.1 LBMM01001166 KMQ96792.1 GL439967 EFN66489.1 KK107119 QOIP01000009 EZA58727.1 RLU18942.1 GL453340 EFN76207.1 KZ288344 PBC27681.1 JR035723 JR035724 AEY56633.1 AEY56634.1 KQ766043 OAD53765.1 NNAY01000939 OXU25822.1 KK852595 KDR20642.1 GBYB01012845 JAG82612.1 KQ414813 KOC60537.1 KQ434874 KZC09650.1 KQ976453 KYM85345.1 KQ982612 KYQ53875.1 GALA01000989 JAA93863.1 GL888206 EGI65178.1 KQ981701 KYN37500.1 KQ978949 KYN27359.1 APCN01004534 APCN01004535 AAAB01008980 EAA43463.4 GFDL01002339 JAV32706.1 GFDL01002340 JAV32705.1 GAPW01002763 JAC10835.1 GAMD01002332 JAA99258.1 CH477692 EAT37221.1 AXCN02000751 AXCN02000752 GL767674 EFZ13266.1 ADTU01008618 ADTU01008619 ADTU01008620 GGFK01002957 MBW36278.1 GGFM01001261 MBW22012.1 GGFL01004447 MBW68625.1 KF813098 AHH35011.1 GGFJ01007600 MBW56741.1 UFQT01000215 SSX21830.1 GEBQ01025457 JAT14520.1 GFDF01009051 JAV05033.1 DS235812 EEB17426.1 KQ977513 KYN02163.1 GECU01030411 JAS77295.1 GFDF01009049 JAV05035.1 GECU01012359 JAS95347.1 GDJX01019345 JAT48591.1 NEVH01002699 PNF41568.1 GBHO01005255 GBRD01000598 GDHC01008674 JAG38349.1 JAG65223.1 JAQ09955.1 PNF41573.1 GEZM01077309 JAV63332.1 CH940654 EDW57958.1 GDKW01001068 JAI55527.1 GAHY01000563 JAA76947.1 KB631641 ERL84900.1 KRF78050.1 KRF78051.1 KF813100 AHH35013.1 CP012523 ALC39250.1 GECL01001960 JAP04164.1 CM000158 EDW89933.1 GBBI01000815 JAC17897.1 CH477641 EAT37924.1 EAT37926.1 BT004839 AAO45195.1 GAMC01015562 GAMC01015561 JAB90993.1 CH916372 EDV98949.1 GEDC01017263 JAS20035.1 CM002910 KMY91349.1 CH480833 EDW46462.1 CH954179 EDV54434.1 AF202633 AE014134 AY314744 BT044501 AAF17587.1 AAF53995.1 AAN11121.1 AAN11122.1 AAP79432.1 ACH95275.1 KRJ98796.1 KRJ98797.1 KRJ98798.1 GFDG01000654 JAV18145.1 EZ422307 ADD18583.1 GAMC01015560 JAB90995.1 KMY91350.1 AGB93210.1 KQS61786.1 CH933807 EDW11967.1 CH964214 EDW80397.1 GBXI01005852 JAD08440.1 AAAB01008964 EAA12801.3 KRG03005.1 KRG03006.1 KRG03007.1 GDHF01015849 GDHF01014106 GDHF01013089 GDHF01004473 GDHF01001608 JAI36465.1 JAI38208.1 JAI39225.1 JAI47841.1 JAI50706.1 EFN66488.1 GAKP01015964 GAKP01015962 JAC42988.1 GBXI01016170 JAC98121.1 CH902620 EDV31871.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002358 UP000036403 UP000000311 UP000005203 UP000053097 UP000279307 UP000008237 UP000242457 UP000215335 UP000027135 UP000053825 UP000076502 UP000078540 UP000075809 UP000007755 UP000078541 UP000078492 UP000075840 UP000007062 UP000075903 UP000008820 UP000075886 UP000005205 UP000075884 UP000009046 UP000078542 UP000076408 UP000235965 UP000008792 UP000030742 UP000092553 UP000002282 UP000001070 UP000001292 UP000008711 UP000000803 UP000192221 UP000009192 UP000075880 UP000076407 UP000007798 UP000007801
UP000002358 UP000036403 UP000000311 UP000005203 UP000053097 UP000279307 UP000008237 UP000242457 UP000215335 UP000027135 UP000053825 UP000076502 UP000078540 UP000075809 UP000007755 UP000078541 UP000078492 UP000075840 UP000007062 UP000075903 UP000008820 UP000075886 UP000005205 UP000075884 UP000009046 UP000078542 UP000076408 UP000235965 UP000008792 UP000030742 UP000092553 UP000002282 UP000001070 UP000001292 UP000008711 UP000000803 UP000192221 UP000009192 UP000075880 UP000076407 UP000007798 UP000007801
Pfam
PF00287 Na_K-ATPase
Interpro
SUPFAM
SSF48225
SSF48225
Gene 3D
ProteinModelPortal
H9J970
A0A2H1VU14
A0A2W1BL15
A0A2A4K154
A0A194RBK0
A0A194PYR3
+ More
A0A212F3C9 A0A0L7LDP8 U5EVF5 K7IVU0 A0A0J7L1W9 E2AJ49 A0A088A4D4 A0A026WRS7 E2C794 A0A2A3E9E4 V9I8J8 A0A310SHK5 A0A232F5D8 A0A067RM12 A0A0C9RVD0 A0A0L7QPH5 A0A154PEL0 A0A195BKU6 A0A151X0X1 T1E275 F4WKT6 A0A151JVT6 A0A151JM06 A0A182IDW3 Q7PK91 A0A1Q3FYT7 A0A1Q3FYQ3 A0A023EPB7 T1E8B1 A0A182V944 Q16RY8 A0A182QDC3 E9J1Q5 A0A158P3Y7 A0A2M4A6M2 A0A2M3Z0F1 A0A2M4CTN4 W5UB64 A0A2M4BUW5 A0A336LVH5 A0A1B6KSW6 A0A182NUX5 A0A1L8DEZ6 E0VVM0 A0A195CN73 A0A1B6HRM5 A0A1L8DF72 A0A1B6J844 A0A182Y5L6 A0A1S4JYY7 A0A1D1Y1U2 A0A2J7RL52 A0A0A9Z1J2 A0A2J7RL67 A0A1Y1KPJ3 B4M9T2 A0A0P4VU23 R4FNJ2 U4U4L8 A0A0Q9VZG3 W5U5U7 A0A0M4EQG5 A0A0V0G820 B4P6W4 A0A023F8Y2 Q16TR9 Q86NM2 W8B1U8 B4JPW9 A0A1B6D385 A0A0J9R6J8 B4IFP0 B3NLW8 Q7JS69 A0A0R1DPA3 A0A1L8EHL9 D3TLK5 W8AQD4 A0A1W4UY15 A0A0J9TSR4 M9PDX8 A0A0Q5VXC3 B4KEZ8 A0A182JE55 A0A2C9H707 B4N7Q8 A0A0A1XBW7 Q7Q3B0 A0A0Q9X4L1 A0A0K8W9K6 E2AJ48 A0A034VN44 A0A0A1WGW0 B3MMB1
A0A212F3C9 A0A0L7LDP8 U5EVF5 K7IVU0 A0A0J7L1W9 E2AJ49 A0A088A4D4 A0A026WRS7 E2C794 A0A2A3E9E4 V9I8J8 A0A310SHK5 A0A232F5D8 A0A067RM12 A0A0C9RVD0 A0A0L7QPH5 A0A154PEL0 A0A195BKU6 A0A151X0X1 T1E275 F4WKT6 A0A151JVT6 A0A151JM06 A0A182IDW3 Q7PK91 A0A1Q3FYT7 A0A1Q3FYQ3 A0A023EPB7 T1E8B1 A0A182V944 Q16RY8 A0A182QDC3 E9J1Q5 A0A158P3Y7 A0A2M4A6M2 A0A2M3Z0F1 A0A2M4CTN4 W5UB64 A0A2M4BUW5 A0A336LVH5 A0A1B6KSW6 A0A182NUX5 A0A1L8DEZ6 E0VVM0 A0A195CN73 A0A1B6HRM5 A0A1L8DF72 A0A1B6J844 A0A182Y5L6 A0A1S4JYY7 A0A1D1Y1U2 A0A2J7RL52 A0A0A9Z1J2 A0A2J7RL67 A0A1Y1KPJ3 B4M9T2 A0A0P4VU23 R4FNJ2 U4U4L8 A0A0Q9VZG3 W5U5U7 A0A0M4EQG5 A0A0V0G820 B4P6W4 A0A023F8Y2 Q16TR9 Q86NM2 W8B1U8 B4JPW9 A0A1B6D385 A0A0J9R6J8 B4IFP0 B3NLW8 Q7JS69 A0A0R1DPA3 A0A1L8EHL9 D3TLK5 W8AQD4 A0A1W4UY15 A0A0J9TSR4 M9PDX8 A0A0Q5VXC3 B4KEZ8 A0A182JE55 A0A2C9H707 B4N7Q8 A0A0A1XBW7 Q7Q3B0 A0A0Q9X4L1 A0A0K8W9K6 E2AJ48 A0A034VN44 A0A0A1WGW0 B3MMB1
PDB
4XE5
E-value=8.55679e-24,
Score=272
Ontologies
GO
PANTHER
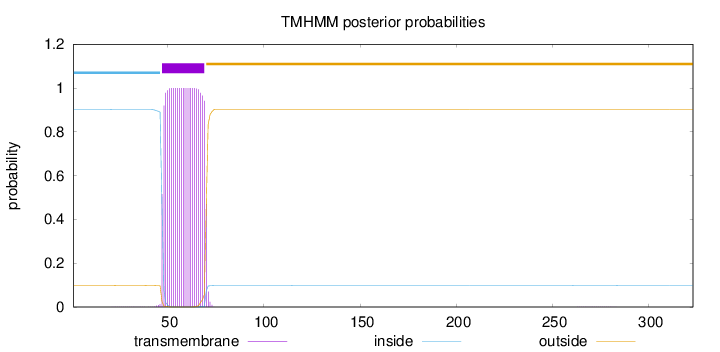
Topology
Length:
323
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89501
Exp number, first 60 AAs:
13.43555
Total prob of N-in:
0.90235
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 69
outside
70 - 323
Population Genetic Test Statistics
Pi
178.237194
Theta
16.140636
Tajima's D
0.019599
CLR
1.291727
CSRT
0.382380880955952
Interpretation
Uncertain
