Gene
KWMTBOMO01632
Pre Gene Modal
BGIBMGA006095
Annotation
PREDICTED:_laminin_subunit_alpha-2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.283 Nuclear Reliability : 1.238 PlasmaMembrane Reliability : 1.5
Sequence
CDS
ATGCTCCAATCCAATATTCGCCTCGTGTTAATTGAACATTATGCAATTTGGTTTGTCGTAGGCCGGGCGGTGCTGGACCGGCTCGCGAGCACGACCGTGAGTGATGTGGACACGGTCACGAGCGCAGCGGTTAGCGGCTTGTTGCCCGCTCCGCTAGACGTCGCGCCGTACTCTGTGATCGCGGCCAATGCGACTTGCGGAGAAGATGGAGAAGAAGAATATTGTCGGGACACGCCGGGAAAGCGTGGAACCGTCTGTGATATCTGCGAGGGTCCGGACGGCTCGTCTTTCCGCAGACATCCGCCATCACACGCACTGGACGGAGACACTTCAACTTGGTGGCAAAGTCCTACATTGGCTGCAGGCGAGCAGTATAAACACGTCGAGTTAATCACAACTTTGCCAGACAAAATGGAACTGCTCCACGTCATCATTAAGTCAGGTCCGTCACCTCGACCCTTGGCCTGGTCGCTCGAGGTATCCGCATCGGAAAGCGGCGAAGACTGGCGCATGATCCGAGCGTATGGAGACAAAGACCACTGCAAGAAAATTTGGGATTTGCGACCAGAGCGACGGAGGAGGAAAGCTAGGGCGGCAAAGAGAATTAATAGAGCTCATAAAATTAGTTGTTCTACACAATTTGCTAATTATAAACCATTAGAAAATGGAGAGATGCATGTGGCTGTCGGCGAAGGAGTATCAGCTCGAAGAGTTCGTATATCGTTTCGCGGCGCTCACCCGACTCCGTCTCGGCAGCAGTACTACACCGTGCGATCCCTGACGTTGGCGGCGAGATGCTTGTGCCATGGACACGCCCAGCGATGTGCTGTCGATAATAAAGGTGCGAAATGCGAATGCTTGCATGCTACGTGTGGATCTCATTGCCATCGATGCTGTTCTGGTGGATCTTGGGCTCCACACAAAATCTGTGATGAGACGCTGGAGTGCCTCTGCGGAGAACGGGGACAGTGCTCTTTCGATGACACTGGCGCTACTCTGTGTCTTAACTGTACGGAAAATCGTGGTGGACCATCATGCGACAAATGTTTATTCGGATATTACAATACGGTGTCGGACGGGCCCTGCGTGCCCTGTGACTGTGACCCTGAAGGATCTGATGGTTCCTGCGAATGGGACAATAAGCGTCACCGAGTATCGTGCAACTGTCTTCCTGGCTTCACCGGTCACCTCTGCGACTCCTGCGAGTCCTCGAACGCTGTTTTTCCTTTATGCCATGACATCCCCACCGAACCGCCGTGCAAATGTGACGTCAGAGGCATCGTGGATCCCACAAGAGAGTGCGATGAAGTCTGTGAATGTAAACTCAACGTGATAGGAGAGCGATGCGACACGTGCGCTTCTGGACACTTTGGGCTCAGCTCTGACCTGAGGGAAGGTTGCAGACCATGCTATTGTTCACACGTTACAGACATCTGCGAGCTCGCTGCACCAGACCCCAACACCCCTCATGATATAGTCCTGCCGCTGGGCGAAGCGTGGATGATCAGCGATTCCATAGCTAACGAGACGCTGGAACCATCACTGGATGAACAAGGGAAACCTTTTGTCATTAGTTACGAGGTAGAAGGGTGGGAGAACTTTTACTGGTTGAGCAGGTCTTTCAACGGTGAACAATTTGATTCGTATGGCGGAGAAATCAGAGCAAGCTTATTCTGGGGCGTAGTGCGTGGTGATTCCGGAGGGAATCCTACCGTTGGTCCTGATGTGATGCTGATTTCTGCTGATGGCAGAATGCTAGCGTACGCAAACACAACCCACGAAACTCCTGGACAGCTGGAGTTATCCGTACCACTGATCGAAGGGAGCTGGTACGTGGTGGGAGATGATGAACCTTGCAGCCGTTTGCAGCTGATGGACGCGTTGCGAGATCTTCGAACTCTCATGATTAAAGCTCACTTTCATATGGACCAGGATGAGATCCGCCTGGAAAACGCTGAAATCCGTGCCCCCGTGAAGCCAGTTAGCGAGGTGTGCTCTTGTCCTGCGGGGTACACTGGAGTCCAGTGCTCATCGTGCGCGTGGTCTCACGCCAGGATCCTCCACGCAGCGGGTGCAGTGCCACCCTTCGAGTGCGTGCCGTGCGCTTGCAATGAACACGCTTCTTGTGATACCGTGGAAGGCCCTTGTGGCTCTTGTCAGCACAACACGACCGGTGCTCACTGCGAGCGCTGTCTTCCCGGCCATTATGGAAATCCAGTCCAGGGTGCTTGCAAGCCGTGCGCCTGTCCATTGTACCTGCCTTCGAATAACTTCAGCCCGAACTGCGCTCTAGCCTCTGCTGAAGGCGACGAGTTCGTCTGCACTCAATGTCCTGACGGCTACACCGGAGACCACTGCGAAAATTGTGATTTCGGGTACTGGGGCTCCCCCACGACACCCGGCGGCTCGTGTCAAGAGTGCGCGTGTGGGGGGTCGCCGTGTCATCCCACTACTGGACTTTGTCTCACTTGTCCCCCGCACACAGAGGGCGCTAGGAAACATCCTGAAGAAGAATTCGTTGAGAGGAATATTGTTAATACCAGATTACAGTGCTCAAATTAG
Protein
MLQSNIRLVLIEHYAIWFVVGRAVLDRLASTTVSDVDTVTSAAVSGLLPAPLDVAPYSVIAANATCGEDGEEEYCRDTPGKRGTVCDICEGPDGSSFRRHPPSHALDGDTSTWWQSPTLAAGEQYKHVELITTLPDKMELLHVIIKSGPSPRPLAWSLEVSASESGEDWRMIRAYGDKDHCKKIWDLRPERRRRKARAAKRINRAHKISCSTQFANYKPLENGEMHVAVGEGVSARRVRISFRGAHPTPSRQQYYTVRSLTLAARCLCHGHAQRCAVDNKGAKCECLHATCGSHCHRCCSGGSWAPHKICDETLECLCGERGQCSFDDTGATLCLNCTENRGGPSCDKCLFGYYNTVSDGPCVPCDCDPEGSDGSCEWDNKRHRVSCNCLPGFTGHLCDSCESSNAVFPLCHDIPTEPPCKCDVRGIVDPTRECDEVCECKLNVIGERCDTCASGHFGLSSDLREGCRPCYCSHVTDICELAAPDPNTPHDIVLPLGEAWMISDSIANETLEPSLDEQGKPFVISYEVEGWENFYWLSRSFNGEQFDSYGGEIRASLFWGVVRGDSGGNPTVGPDVMLISADGRMLAYANTTHETPGQLELSVPLIEGSWYVVGDDEPCSRLQLMDALRDLRTLMIKAHFHMDQDEIRLENAEIRAPVKPVSEVCSCPAGYTGVQCSSCAWSHARILHAAGAVPPFECVPCACNEHASCDTVEGPCGSCQHNTTGAHCERCLPGHYGNPVQGACKPCACPLYLPSNNFSPNCALASAEGDEFVCTQCPDGYTGDHCENCDFGYWGSPTTPGGSCQECACGGSPCHPTTGLCLTCPPHTEGARKHPEEEFVERNIVNTRLQCSN
Summary
Uniprot
A0A2H1WQI3
A0A194PYQ8
A0A212F7W4
A0A194R6J1
A0A2A4K892
A0A088AQK5
+ More
A0A0L7RE06 A0A026WQX0 A0A151I948 A0A154NX77 A0A151WKC6 A0A195EW19 A0A151IWU9 Q7PN80 A0A1B0CBA8 A0A195BRX4 A0A139WHZ5 D6WJN1 A0A1S4GWE4 A0A1S4GX69 A0A084W3W6 A0A0Q9VZ62 B4M8R6 B4KEJ3 R9PY40 Q8IP51 Q9XZC9 A0A067RR33 B4P0J7 A0A0R1DLB2 A0A0J9TN74 A0A3L8DP83 B4JD84 B3MMR4 B3N9T6 A0A0R3NUL1 Q29KB2 A0A3B0KA09 A0A158NNY9 B4GX61 B4MVL9 A0A1I8N927 A0A0M4EG82 A0A1W4UXV6 A0A1W4UWG4 A0A0L0BZ21 F4WV90 A0A2H8TJH9 E2C7R0 A0A2S2PRC5 A0A182IEL8 A0A0A1X1L6 A0A182P0I5 A0A182WTG9 A0A182R3A2 A0A182JQF6 A0A182NLL5 B0WC09 A0A182QPC7 E0VR72 A0A1A9YRJ5 A0A182J6Z0 A0A182YPN5 E2AY88 A0A0A0A754 A0A1B0BIW5 A0A182M890 A0A2J7Q648 Q17L45 F1MMT2 A0A2Y9SLJ5 A0A091IVC3 A0A1B0FLF3 A0A091KA07 F6R9F5 A0A087R6F4 A0A091MR07 M3YT51 A0A336KY12 A0A340WP85 A0A340WUM1 A0A384C4V3 A0A2U4CNF2 A0A085MGE7 A0A1A9W0D4 H0VQL4 A0A384AU28 A0A384AUT2 A0A287A0X6 A0A341CJK3 A0A1D1VGG6 A0A287AP44 A0A087WRP2 A0A384AU19 A0A1D1VDZ8 A0A091GU45 F1M614
A0A0L7RE06 A0A026WQX0 A0A151I948 A0A154NX77 A0A151WKC6 A0A195EW19 A0A151IWU9 Q7PN80 A0A1B0CBA8 A0A195BRX4 A0A139WHZ5 D6WJN1 A0A1S4GWE4 A0A1S4GX69 A0A084W3W6 A0A0Q9VZ62 B4M8R6 B4KEJ3 R9PY40 Q8IP51 Q9XZC9 A0A067RR33 B4P0J7 A0A0R1DLB2 A0A0J9TN74 A0A3L8DP83 B4JD84 B3MMR4 B3N9T6 A0A0R3NUL1 Q29KB2 A0A3B0KA09 A0A158NNY9 B4GX61 B4MVL9 A0A1I8N927 A0A0M4EG82 A0A1W4UXV6 A0A1W4UWG4 A0A0L0BZ21 F4WV90 A0A2H8TJH9 E2C7R0 A0A2S2PRC5 A0A182IEL8 A0A0A1X1L6 A0A182P0I5 A0A182WTG9 A0A182R3A2 A0A182JQF6 A0A182NLL5 B0WC09 A0A182QPC7 E0VR72 A0A1A9YRJ5 A0A182J6Z0 A0A182YPN5 E2AY88 A0A0A0A754 A0A1B0BIW5 A0A182M890 A0A2J7Q648 Q17L45 F1MMT2 A0A2Y9SLJ5 A0A091IVC3 A0A1B0FLF3 A0A091KA07 F6R9F5 A0A087R6F4 A0A091MR07 M3YT51 A0A336KY12 A0A340WP85 A0A340WUM1 A0A384C4V3 A0A2U4CNF2 A0A085MGE7 A0A1A9W0D4 H0VQL4 A0A384AU28 A0A384AUT2 A0A287A0X6 A0A341CJK3 A0A1D1VGG6 A0A287AP44 A0A087WRP2 A0A384AU19 A0A1D1VDZ8 A0A091GU45 F1M614
Pubmed
26354079
22118469
24508170
12364791
18362917
19820115
+ More
24438588 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10189378 24845553 17550304 22936249 30249741 15632085 21347285 25315136 26108605 21719571 20798317 25830018 20566863 25244985 17510324 19393038 19892987 24929829 21993624 27649274 19468303 21183079 15057822
24438588 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10189378 24845553 17550304 22936249 30249741 15632085 21347285 25315136 26108605 21719571 20798317 25830018 20566863 25244985 17510324 19393038 19892987 24929829 21993624 27649274 19468303 21183079 15057822
EMBL
ODYU01010288
SOQ55288.1
KQ459584
KPI98472.1
AGBW02009824
OWR49820.1
+ More
KQ460644 KPJ13282.1 NWSH01000047 PCG80226.1 KQ414611 KOC69212.1 KK107128 EZA58358.1 KQ978303 KYM95327.1 KQ434777 KZC04269.1 KQ983031 KYQ48135.1 KQ981953 KYN32433.1 KQ980843 KYN12309.1 AAAB01008964 EAA12624.5 AJWK01004935 AJWK01004936 AJWK01004937 AJWK01004938 KQ976417 KYM89765.1 KQ971343 KYB27445.1 EFA04469.2 ATLV01020177 ATLV01020178 ATLV01020179 ATLV01020180 ATLV01020181 ATLV01020182 ATLV01020183 ATLV01020184 ATLV01020185 ATLV01020186 KE525295 KFB44910.1 CH940654 KRF77830.1 EDW57592.1 CH933807 EDW11872.2 AE014134 AAF53381.3 AAN10875.1 AF135118 AAD31714.1 KK852514 KDR22194.1 CM000157 EDW88991.2 KRJ98117.1 CM002910 KMY90230.1 QOIP01000006 RLU21689.1 CH916368 EDW03257.1 CH902620 EDV31955.2 CH954177 EDV58581.1 CH379061 KRT04865.1 EAL33263.2 OUUW01000006 SPP82486.1 ADTU01021838 ADTU01021839 ADTU01021840 ADTU01021841 ADTU01021842 ADTU01021843 ADTU01021844 ADTU01021845 ADTU01021846 ADTU01021847 CH479195 EDW27388.1 CH963857 EDW75739.1 CP012523 ALC39642.1 JRES01001125 KNC25302.1 GL888384 EGI61856.1 GFXV01002027 MBW13832.1 GL453395 EFN76043.1 GGMR01019308 MBY31927.1 APCN01005209 GBXI01009637 JAD04655.1 DS231884 EDS43069.1 AXCN02000611 AXCN02000612 DS235459 EEB15878.1 GL443772 EFN61600.1 KL870806 KGL89293.1 JXJN01015211 AXCM01010359 NEVH01017541 PNF24055.1 CH477219 EAT47381.1 KL218257 KFP03591.1 CCAG010017313 KK502094 KFP20586.1 KL226140 KFM09058.1 KK835858 KFP79923.1 AEYP01006796 AEYP01006797 AEYP01006798 AEYP01006799 AEYP01006800 AEYP01006801 AEYP01006802 AEYP01006803 AEYP01006804 AEYP01006805 UFQS01001005 UFQT01001005 SSX08493.1 SSX28409.1 KL363194 KFD56293.1 AAKN02038162 AAKN02038163 AAKN02038164 AAKN02038165 AAKN02038166 AAKN02038167 AAKN02038168 AAKN02038169 AAKN02038170 AAKN02038171 AEMK02000001 BDGG01000005 GAU99875.1 AC101709 AC152982 AC153800 AC155942 AC167232 AC171406 GAU99874.1 KL447830 KFO77647.1 AABR07000583 AABR07000584 AABR07000585 AABR07000586 AABR07000587
KQ460644 KPJ13282.1 NWSH01000047 PCG80226.1 KQ414611 KOC69212.1 KK107128 EZA58358.1 KQ978303 KYM95327.1 KQ434777 KZC04269.1 KQ983031 KYQ48135.1 KQ981953 KYN32433.1 KQ980843 KYN12309.1 AAAB01008964 EAA12624.5 AJWK01004935 AJWK01004936 AJWK01004937 AJWK01004938 KQ976417 KYM89765.1 KQ971343 KYB27445.1 EFA04469.2 ATLV01020177 ATLV01020178 ATLV01020179 ATLV01020180 ATLV01020181 ATLV01020182 ATLV01020183 ATLV01020184 ATLV01020185 ATLV01020186 KE525295 KFB44910.1 CH940654 KRF77830.1 EDW57592.1 CH933807 EDW11872.2 AE014134 AAF53381.3 AAN10875.1 AF135118 AAD31714.1 KK852514 KDR22194.1 CM000157 EDW88991.2 KRJ98117.1 CM002910 KMY90230.1 QOIP01000006 RLU21689.1 CH916368 EDW03257.1 CH902620 EDV31955.2 CH954177 EDV58581.1 CH379061 KRT04865.1 EAL33263.2 OUUW01000006 SPP82486.1 ADTU01021838 ADTU01021839 ADTU01021840 ADTU01021841 ADTU01021842 ADTU01021843 ADTU01021844 ADTU01021845 ADTU01021846 ADTU01021847 CH479195 EDW27388.1 CH963857 EDW75739.1 CP012523 ALC39642.1 JRES01001125 KNC25302.1 GL888384 EGI61856.1 GFXV01002027 MBW13832.1 GL453395 EFN76043.1 GGMR01019308 MBY31927.1 APCN01005209 GBXI01009637 JAD04655.1 DS231884 EDS43069.1 AXCN02000611 AXCN02000612 DS235459 EEB15878.1 GL443772 EFN61600.1 KL870806 KGL89293.1 JXJN01015211 AXCM01010359 NEVH01017541 PNF24055.1 CH477219 EAT47381.1 KL218257 KFP03591.1 CCAG010017313 KK502094 KFP20586.1 KL226140 KFM09058.1 KK835858 KFP79923.1 AEYP01006796 AEYP01006797 AEYP01006798 AEYP01006799 AEYP01006800 AEYP01006801 AEYP01006802 AEYP01006803 AEYP01006804 AEYP01006805 UFQS01001005 UFQT01001005 SSX08493.1 SSX28409.1 KL363194 KFD56293.1 AAKN02038162 AAKN02038163 AAKN02038164 AAKN02038165 AAKN02038166 AAKN02038167 AAKN02038168 AAKN02038169 AAKN02038170 AAKN02038171 AEMK02000001 BDGG01000005 GAU99875.1 AC101709 AC152982 AC153800 AC155942 AC167232 AC171406 GAU99874.1 KL447830 KFO77647.1 AABR07000583 AABR07000584 AABR07000585 AABR07000586 AABR07000587
Proteomes
UP000053268
UP000007151
UP000053240
UP000218220
UP000005203
UP000053825
+ More
UP000053097 UP000078542 UP000076502 UP000075809 UP000078541 UP000078492 UP000007062 UP000092461 UP000078540 UP000007266 UP000030765 UP000008792 UP000009192 UP000000803 UP000027135 UP000002282 UP000279307 UP000001070 UP000007801 UP000008711 UP000001819 UP000268350 UP000005205 UP000008744 UP000007798 UP000095301 UP000092553 UP000192221 UP000037069 UP000007755 UP000008237 UP000075840 UP000075885 UP000076407 UP000075900 UP000075881 UP000075884 UP000002320 UP000075886 UP000009046 UP000092443 UP000075880 UP000076408 UP000000311 UP000053858 UP000092460 UP000075883 UP000235965 UP000008820 UP000009136 UP000248484 UP000054308 UP000092444 UP000053119 UP000002281 UP000053286 UP000000715 UP000265300 UP000261680 UP000245320 UP000091820 UP000005447 UP000261681 UP000008227 UP000252040 UP000186922 UP000000589 UP000053760 UP000002494
UP000053097 UP000078542 UP000076502 UP000075809 UP000078541 UP000078492 UP000007062 UP000092461 UP000078540 UP000007266 UP000030765 UP000008792 UP000009192 UP000000803 UP000027135 UP000002282 UP000279307 UP000001070 UP000007801 UP000008711 UP000001819 UP000268350 UP000005205 UP000008744 UP000007798 UP000095301 UP000092553 UP000192221 UP000037069 UP000007755 UP000008237 UP000075840 UP000075885 UP000076407 UP000075900 UP000075881 UP000075884 UP000002320 UP000075886 UP000009046 UP000092443 UP000075880 UP000076408 UP000000311 UP000053858 UP000092460 UP000075883 UP000235965 UP000008820 UP000009136 UP000248484 UP000054308 UP000092444 UP000053119 UP000002281 UP000053286 UP000000715 UP000265300 UP000261680 UP000245320 UP000091820 UP000005447 UP000261681 UP000008227 UP000252040 UP000186922 UP000000589 UP000053760 UP000002494
Pfam
Interpro
IPR013032
EGF-like_CS
+ More
IPR008211 Laminin_N
IPR000742 EGF-like_dom
IPR038684 Laminin_N_sf
IPR002049 Laminin_EGF
IPR000034 Laminin_IV
IPR001190 SRCR
IPR008979 Galactose-bd-like_sf
IPR013087 Znf_C2H2_type
IPR013320 ConA-like_dom_sf
IPR001791 Laminin_G
IPR009254 Laminin_aI
IPR009030 Growth_fac_rcpt_cys_sf
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR010307 Laminin_dom_II
IPR000679 Znf_GATA
IPR008211 Laminin_N
IPR000742 EGF-like_dom
IPR038684 Laminin_N_sf
IPR002049 Laminin_EGF
IPR000034 Laminin_IV
IPR001190 SRCR
IPR008979 Galactose-bd-like_sf
IPR013087 Znf_C2H2_type
IPR013320 ConA-like_dom_sf
IPR001791 Laminin_G
IPR009254 Laminin_aI
IPR009030 Growth_fac_rcpt_cys_sf
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR010307 Laminin_dom_II
IPR000679 Znf_GATA
Gene 3D
ProteinModelPortal
A0A2H1WQI3
A0A194PYQ8
A0A212F7W4
A0A194R6J1
A0A2A4K892
A0A088AQK5
+ More
A0A0L7RE06 A0A026WQX0 A0A151I948 A0A154NX77 A0A151WKC6 A0A195EW19 A0A151IWU9 Q7PN80 A0A1B0CBA8 A0A195BRX4 A0A139WHZ5 D6WJN1 A0A1S4GWE4 A0A1S4GX69 A0A084W3W6 A0A0Q9VZ62 B4M8R6 B4KEJ3 R9PY40 Q8IP51 Q9XZC9 A0A067RR33 B4P0J7 A0A0R1DLB2 A0A0J9TN74 A0A3L8DP83 B4JD84 B3MMR4 B3N9T6 A0A0R3NUL1 Q29KB2 A0A3B0KA09 A0A158NNY9 B4GX61 B4MVL9 A0A1I8N927 A0A0M4EG82 A0A1W4UXV6 A0A1W4UWG4 A0A0L0BZ21 F4WV90 A0A2H8TJH9 E2C7R0 A0A2S2PRC5 A0A182IEL8 A0A0A1X1L6 A0A182P0I5 A0A182WTG9 A0A182R3A2 A0A182JQF6 A0A182NLL5 B0WC09 A0A182QPC7 E0VR72 A0A1A9YRJ5 A0A182J6Z0 A0A182YPN5 E2AY88 A0A0A0A754 A0A1B0BIW5 A0A182M890 A0A2J7Q648 Q17L45 F1MMT2 A0A2Y9SLJ5 A0A091IVC3 A0A1B0FLF3 A0A091KA07 F6R9F5 A0A087R6F4 A0A091MR07 M3YT51 A0A336KY12 A0A340WP85 A0A340WUM1 A0A384C4V3 A0A2U4CNF2 A0A085MGE7 A0A1A9W0D4 H0VQL4 A0A384AU28 A0A384AUT2 A0A287A0X6 A0A341CJK3 A0A1D1VGG6 A0A287AP44 A0A087WRP2 A0A384AU19 A0A1D1VDZ8 A0A091GU45 F1M614
A0A0L7RE06 A0A026WQX0 A0A151I948 A0A154NX77 A0A151WKC6 A0A195EW19 A0A151IWU9 Q7PN80 A0A1B0CBA8 A0A195BRX4 A0A139WHZ5 D6WJN1 A0A1S4GWE4 A0A1S4GX69 A0A084W3W6 A0A0Q9VZ62 B4M8R6 B4KEJ3 R9PY40 Q8IP51 Q9XZC9 A0A067RR33 B4P0J7 A0A0R1DLB2 A0A0J9TN74 A0A3L8DP83 B4JD84 B3MMR4 B3N9T6 A0A0R3NUL1 Q29KB2 A0A3B0KA09 A0A158NNY9 B4GX61 B4MVL9 A0A1I8N927 A0A0M4EG82 A0A1W4UXV6 A0A1W4UWG4 A0A0L0BZ21 F4WV90 A0A2H8TJH9 E2C7R0 A0A2S2PRC5 A0A182IEL8 A0A0A1X1L6 A0A182P0I5 A0A182WTG9 A0A182R3A2 A0A182JQF6 A0A182NLL5 B0WC09 A0A182QPC7 E0VR72 A0A1A9YRJ5 A0A182J6Z0 A0A182YPN5 E2AY88 A0A0A0A754 A0A1B0BIW5 A0A182M890 A0A2J7Q648 Q17L45 F1MMT2 A0A2Y9SLJ5 A0A091IVC3 A0A1B0FLF3 A0A091KA07 F6R9F5 A0A087R6F4 A0A091MR07 M3YT51 A0A336KY12 A0A340WP85 A0A340WUM1 A0A384C4V3 A0A2U4CNF2 A0A085MGE7 A0A1A9W0D4 H0VQL4 A0A384AU28 A0A384AUT2 A0A287A0X6 A0A341CJK3 A0A1D1VGG6 A0A287AP44 A0A087WRP2 A0A384AU19 A0A1D1VDZ8 A0A091GU45 F1M614
PDB
2Y38
E-value=1.96728e-21,
Score=256
Ontologies
GO
GO:0016020
GO:0005044
GO:0003676
GO:0016021
GO:0030334
GO:0045995
GO:0030155
GO:0005102
GO:0009888
GO:0007411
GO:0009887
GO:0005604
GO:0008406
GO:0007502
GO:0005201
GO:0007155
GO:0043083
GO:0032224
GO:0042383
GO:0031594
GO:0008270
GO:0043565
GO:0006355
GO:0014037
GO:0043197
GO:0005515
GO:0003824
GO:0004190
GO:0006508
GO:0006811
GO:0003779
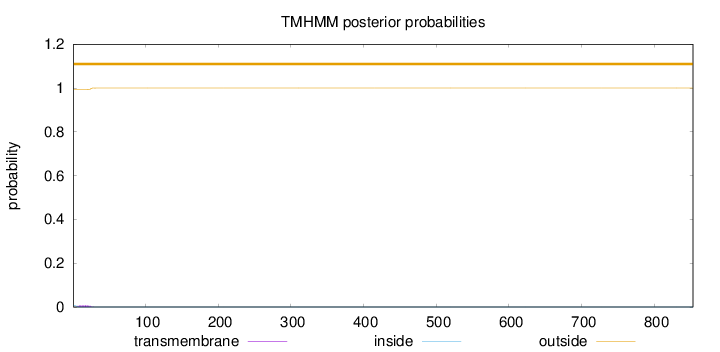
Topology
Length:
853
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11693
Exp number, first 60 AAs:
0.11687
Total prob of N-in:
0.00612
outside
1 - 853
Population Genetic Test Statistics
Pi
234.154969
Theta
175.956624
Tajima's D
0.887532
CLR
0.35963
CSRT
0.630268486575671
Interpretation
Uncertain
