Gene
KWMTBOMO01631
Annotation
PREDICTED:_uncharacterized_protein_LOC101745917_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.552
Sequence
CDS
ATGGAATCACTATGCAAGGACCACTTTCTTCAACGTCAATACAGAAAGTTAGACGGAGGTGTCCTGTGGTCAAGGAACGAGAGGAATCTGGACTGCACCGTGACTTTCCAGACGCACAGCATCCTCCAGAGGTTCATGCTGCATTTCGATCTACTGCAATTGGATTGCAATGACCATCTCTACGTGTACGACGGGGCGCATGCCACCGCGCCGCCGAAGGCTGACTTATCGTGCCGGAACACCAAGCAGCAGGTCGGAGCCCTGTTCACGCGCAGCAACTTCGTGACCCTGAAGTACGTCACTGACAACTGGGGCACGGACGCGAACGGCTTCAAGCTCGTCATCACTGCAGTCAAAGATCCTAAGCACGGCTGCAAGGAGTTCAGGTGCAAGCAGAGAGAGTTCTGCGTGAGTGCAGATCTAACCTGCGACGGCGTGGACCACTGCGCCGATGGTTCAGATGAAGACACGGTCGCTCTATGCCCAGAGAGCGGCGGTAGCGGTTCGAGCAGCACGTGGTTGGTGGTGGCGGGAGGGGCCGGGCTGTTGCTGGCGCTGATCGCCGCGACCCTCGGCTGCTGTCTCTGCCGACGCCGCGCTGCCAACCGCCGCACGCATCTACATTGTTAG
Protein
MESLCKDHFLQRQYRKLDGGVLWSRNERNLDCTVTFQTHSILQRFMLHFDLLQLDCNDHLYVYDGAHATAPPKADLSCRNTKQQVGALFTRSNFVTLKYVTDNWGTDANGFKLVITAVKDPKHGCKEFRCKQREFCVSADLTCDGVDHCADGSDEDTVALCPESGGSGSSSTWLVVAGGAGLLLALIAATLGCCLCRRRAANRRTHLHC
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A2H1W0V7
A0A2A4K8V2
A0A2W1BQD6
A0A212F7V5
A0A194R7S0
A0A1W4WHR7
+ More
N6UJA9 U4U0N2 W8BH29 D6WJN0 A0A336LKG5 A0A0K8UNH7 A0A0M4E9N4 B4LCS1 A0A0A1WKL1 A0A1B6GYB6 B4KUT5 Q29DX5 B4IZB1 B4H489 B4N4K8 A0A3B0JVK1 A0A0J9RTW4 Q5BIG2 B3NH00 B4PFN7 A0A1I8N168 A0A1B6DDM6 A0A1B6M420 B4HEY3 A0A1W4W7A4 B3M4B9 A0A0L0CCC9 A0A232F946 A0A182JBS0 Q7Q067 A0A182MYJ0 K7J1N4 A0A1B0G277 A0A182XD76 A0A182HQ20 A0A182K277 A0A182PBQ6 A0A067REC2 A0A182FU37 A0A158NNW2 A0A182W971 A0A182SCI2 A0A0L7REG0 A0A182R5F1 A0A0K8SEC3 A0A0K8SED0 A0A1B0CL85 A0A1J1HGP5 X1WIY0 A0A2S2NWN5 A0A2S2R5J2 A0A0P5C2J9 A0A0P5C2V6 A0A0P4XXH3 E9G466 A0A0P5J0H7 A0A0P5KKF5 A0A0P4XG62 A0A0P6JKJ8 A0A1J1HSY6 A0A182Y1T9 A0A3Q0IU93 A0A0P5CXW4 A0A2R5LHX7 A0A087UFI0 A0A293LPM2 V5GV24 A0A1B0B1B3 A0A0K2UVK0 B7QHM6 A0A0K2UW67 A0A023GAS5 A0A1E1XVG0 A0A1A9YEM1 A0A1V9XLZ5 A0A131Z3M2 A0A2P6L132 A0A0C9SEB1 T1JS33 T1J4X9
N6UJA9 U4U0N2 W8BH29 D6WJN0 A0A336LKG5 A0A0K8UNH7 A0A0M4E9N4 B4LCS1 A0A0A1WKL1 A0A1B6GYB6 B4KUT5 Q29DX5 B4IZB1 B4H489 B4N4K8 A0A3B0JVK1 A0A0J9RTW4 Q5BIG2 B3NH00 B4PFN7 A0A1I8N168 A0A1B6DDM6 A0A1B6M420 B4HEY3 A0A1W4W7A4 B3M4B9 A0A0L0CCC9 A0A232F946 A0A182JBS0 Q7Q067 A0A182MYJ0 K7J1N4 A0A1B0G277 A0A182XD76 A0A182HQ20 A0A182K277 A0A182PBQ6 A0A067REC2 A0A182FU37 A0A158NNW2 A0A182W971 A0A182SCI2 A0A0L7REG0 A0A182R5F1 A0A0K8SEC3 A0A0K8SED0 A0A1B0CL85 A0A1J1HGP5 X1WIY0 A0A2S2NWN5 A0A2S2R5J2 A0A0P5C2J9 A0A0P5C2V6 A0A0P4XXH3 E9G466 A0A0P5J0H7 A0A0P5KKF5 A0A0P4XG62 A0A0P6JKJ8 A0A1J1HSY6 A0A182Y1T9 A0A3Q0IU93 A0A0P5CXW4 A0A2R5LHX7 A0A087UFI0 A0A293LPM2 V5GV24 A0A1B0B1B3 A0A0K2UVK0 B7QHM6 A0A0K2UW67 A0A023GAS5 A0A1E1XVG0 A0A1A9YEM1 A0A1V9XLZ5 A0A131Z3M2 A0A2P6L132 A0A0C9SEB1 T1JS33 T1J4X9
Pubmed
28756777
22118469
26354079
23537049
24495485
18362917
+ More
19820115 17994087 18057021 25830018 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25315136 26108605 28648823 12364791 20075255 24845553 21347285 21292972 25244985 25765539 29209593 28327890 26830274 26131772
19820115 17994087 18057021 25830018 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25315136 26108605 28648823 12364791 20075255 24845553 21347285 21292972 25244985 25765539 29209593 28327890 26830274 26131772
EMBL
ODYU01005658
SOQ46740.1
NWSH01000047
PCG80223.1
KZ149934
PZC77208.1
+ More
AGBW02009824 OWR49824.1 KQ460644 KPJ13285.1 APGK01022058 APGK01022059 APGK01022060 APGK01022061 KB740396 KB631758 ENN80751.1 ERL85836.1 KB630443 ERL83605.1 GAMC01017616 JAB88939.1 KQ971343 EFA04468.2 UFQS01000028 UFQS01000132 UFQT01000028 UFQT01000132 SSW97799.1 SSX00247.1 SSX18185.1 GDHF01024158 JAI28156.1 CP012525 ALC43841.1 CH940647 EDW70963.2 KRF85214.1 GBXI01015369 JAC98922.1 GECZ01002355 JAS67414.1 CH933809 EDW19341.2 CH379070 EAL30288.3 KRT08792.1 CH916366 EDV96666.1 CH479208 EDW31204.1 CH964095 EDW79082.2 OUUW01000002 SPP76761.1 CM002912 KMY99061.1 KMY99062.1 KMY99063.1 BT021262 AE014296 AAX33410.1 ABW08526.1 CH954178 EDV51457.2 KQS43815.1 CM000159 EDW94186.2 KRK01696.1 GEDC01013603 GEDC01002326 JAS23695.1 JAS34972.1 GEBQ01009312 JAT30665.1 CH480815 EDW41152.1 CH902618 EDV39389.2 KPU78030.1 JRES01000610 KNC29905.1 NNAY01000616 OXU27374.1 AXCP01006766 AAAB01008986 EAA00237.4 CCAG010006679 APCN01003288 KK852514 KDR22196.1 ADTU01021825 ADTU01021826 ADTU01021827 ADTU01021828 ADTU01021829 ADTU01021830 ADTU01021831 ADTU01021832 ADTU01021833 ADTU01021834 KQ414611 KOC69215.1 GBRD01014179 JAG51647.1 GBRD01014180 JAG51646.1 AJWK01017032 CVRI01000004 CRK87183.1 ABLF02007635 ABLF02007636 ABLF02007637 ABLF02020646 ABLF02020647 ABLF02020648 GGMR01008507 MBY21126.1 GGMS01016035 MBY85238.1 GDIP01176455 JAJ46947.1 GDIP01176502 JAJ46900.1 GDIP01235439 JAI87962.1 GL732531 EFX85701.1 GDIQ01208597 JAK43128.1 GDIQ01189978 JAK61747.1 GDIP01242258 JAI81143.1 GDIQ01016222 JAN78515.1 CVRI01000020 CRK91107.1 GDIP01163896 JAJ59506.1 GGLE01004996 MBY09122.1 KK119593 KFM76119.1 GFWV01004050 MAA28780.1 GANP01010213 JAB74255.1 JXJN01007035 JXJN01007036 JXJN01007037 HACA01024566 CDW41927.1 ABJB010923687 ABJB011080025 DS940368 EEC18348.1 HACA01024565 CDW41926.1 GBBM01004451 JAC30967.1 GFAA01000365 JAU03070.1 MNPL01007913 OQR74481.1 GEDV01002563 JAP85994.1 MWRG01002755 PRD32279.1 GBZX01000213 JAG92527.1 CAEY01000458 JH431850
AGBW02009824 OWR49824.1 KQ460644 KPJ13285.1 APGK01022058 APGK01022059 APGK01022060 APGK01022061 KB740396 KB631758 ENN80751.1 ERL85836.1 KB630443 ERL83605.1 GAMC01017616 JAB88939.1 KQ971343 EFA04468.2 UFQS01000028 UFQS01000132 UFQT01000028 UFQT01000132 SSW97799.1 SSX00247.1 SSX18185.1 GDHF01024158 JAI28156.1 CP012525 ALC43841.1 CH940647 EDW70963.2 KRF85214.1 GBXI01015369 JAC98922.1 GECZ01002355 JAS67414.1 CH933809 EDW19341.2 CH379070 EAL30288.3 KRT08792.1 CH916366 EDV96666.1 CH479208 EDW31204.1 CH964095 EDW79082.2 OUUW01000002 SPP76761.1 CM002912 KMY99061.1 KMY99062.1 KMY99063.1 BT021262 AE014296 AAX33410.1 ABW08526.1 CH954178 EDV51457.2 KQS43815.1 CM000159 EDW94186.2 KRK01696.1 GEDC01013603 GEDC01002326 JAS23695.1 JAS34972.1 GEBQ01009312 JAT30665.1 CH480815 EDW41152.1 CH902618 EDV39389.2 KPU78030.1 JRES01000610 KNC29905.1 NNAY01000616 OXU27374.1 AXCP01006766 AAAB01008986 EAA00237.4 CCAG010006679 APCN01003288 KK852514 KDR22196.1 ADTU01021825 ADTU01021826 ADTU01021827 ADTU01021828 ADTU01021829 ADTU01021830 ADTU01021831 ADTU01021832 ADTU01021833 ADTU01021834 KQ414611 KOC69215.1 GBRD01014179 JAG51647.1 GBRD01014180 JAG51646.1 AJWK01017032 CVRI01000004 CRK87183.1 ABLF02007635 ABLF02007636 ABLF02007637 ABLF02020646 ABLF02020647 ABLF02020648 GGMR01008507 MBY21126.1 GGMS01016035 MBY85238.1 GDIP01176455 JAJ46947.1 GDIP01176502 JAJ46900.1 GDIP01235439 JAI87962.1 GL732531 EFX85701.1 GDIQ01208597 JAK43128.1 GDIQ01189978 JAK61747.1 GDIP01242258 JAI81143.1 GDIQ01016222 JAN78515.1 CVRI01000020 CRK91107.1 GDIP01163896 JAJ59506.1 GGLE01004996 MBY09122.1 KK119593 KFM76119.1 GFWV01004050 MAA28780.1 GANP01010213 JAB74255.1 JXJN01007035 JXJN01007036 JXJN01007037 HACA01024566 CDW41927.1 ABJB010923687 ABJB011080025 DS940368 EEC18348.1 HACA01024565 CDW41926.1 GBBM01004451 JAC30967.1 GFAA01000365 JAU03070.1 MNPL01007913 OQR74481.1 GEDV01002563 JAP85994.1 MWRG01002755 PRD32279.1 GBZX01000213 JAG92527.1 CAEY01000458 JH431850
Proteomes
UP000218220
UP000007151
UP000053240
UP000192223
UP000019118
UP000030742
+ More
UP000007266 UP000092553 UP000008792 UP000009192 UP000001819 UP000001070 UP000008744 UP000007798 UP000268350 UP000000803 UP000008711 UP000002282 UP000095301 UP000001292 UP000192221 UP000007801 UP000037069 UP000215335 UP000075880 UP000007062 UP000075884 UP000002358 UP000092444 UP000076407 UP000075840 UP000075881 UP000075885 UP000027135 UP000069272 UP000005205 UP000075920 UP000075901 UP000053825 UP000075900 UP000092461 UP000183832 UP000007819 UP000000305 UP000076408 UP000079169 UP000054359 UP000092460 UP000001555 UP000092443 UP000192247 UP000015104
UP000007266 UP000092553 UP000008792 UP000009192 UP000001819 UP000001070 UP000008744 UP000007798 UP000268350 UP000000803 UP000008711 UP000002282 UP000095301 UP000001292 UP000192221 UP000007801 UP000037069 UP000215335 UP000075880 UP000007062 UP000075884 UP000002358 UP000092444 UP000076407 UP000075840 UP000075881 UP000075885 UP000027135 UP000069272 UP000005205 UP000075920 UP000075901 UP000053825 UP000075900 UP000092461 UP000183832 UP000007819 UP000000305 UP000076408 UP000079169 UP000054359 UP000092460 UP000001555 UP000092443 UP000192247 UP000015104
PRIDE
Interpro
ProteinModelPortal
A0A2H1W0V7
A0A2A4K8V2
A0A2W1BQD6
A0A212F7V5
A0A194R7S0
A0A1W4WHR7
+ More
N6UJA9 U4U0N2 W8BH29 D6WJN0 A0A336LKG5 A0A0K8UNH7 A0A0M4E9N4 B4LCS1 A0A0A1WKL1 A0A1B6GYB6 B4KUT5 Q29DX5 B4IZB1 B4H489 B4N4K8 A0A3B0JVK1 A0A0J9RTW4 Q5BIG2 B3NH00 B4PFN7 A0A1I8N168 A0A1B6DDM6 A0A1B6M420 B4HEY3 A0A1W4W7A4 B3M4B9 A0A0L0CCC9 A0A232F946 A0A182JBS0 Q7Q067 A0A182MYJ0 K7J1N4 A0A1B0G277 A0A182XD76 A0A182HQ20 A0A182K277 A0A182PBQ6 A0A067REC2 A0A182FU37 A0A158NNW2 A0A182W971 A0A182SCI2 A0A0L7REG0 A0A182R5F1 A0A0K8SEC3 A0A0K8SED0 A0A1B0CL85 A0A1J1HGP5 X1WIY0 A0A2S2NWN5 A0A2S2R5J2 A0A0P5C2J9 A0A0P5C2V6 A0A0P4XXH3 E9G466 A0A0P5J0H7 A0A0P5KKF5 A0A0P4XG62 A0A0P6JKJ8 A0A1J1HSY6 A0A182Y1T9 A0A3Q0IU93 A0A0P5CXW4 A0A2R5LHX7 A0A087UFI0 A0A293LPM2 V5GV24 A0A1B0B1B3 A0A0K2UVK0 B7QHM6 A0A0K2UW67 A0A023GAS5 A0A1E1XVG0 A0A1A9YEM1 A0A1V9XLZ5 A0A131Z3M2 A0A2P6L132 A0A0C9SEB1 T1JS33 T1J4X9
N6UJA9 U4U0N2 W8BH29 D6WJN0 A0A336LKG5 A0A0K8UNH7 A0A0M4E9N4 B4LCS1 A0A0A1WKL1 A0A1B6GYB6 B4KUT5 Q29DX5 B4IZB1 B4H489 B4N4K8 A0A3B0JVK1 A0A0J9RTW4 Q5BIG2 B3NH00 B4PFN7 A0A1I8N168 A0A1B6DDM6 A0A1B6M420 B4HEY3 A0A1W4W7A4 B3M4B9 A0A0L0CCC9 A0A232F946 A0A182JBS0 Q7Q067 A0A182MYJ0 K7J1N4 A0A1B0G277 A0A182XD76 A0A182HQ20 A0A182K277 A0A182PBQ6 A0A067REC2 A0A182FU37 A0A158NNW2 A0A182W971 A0A182SCI2 A0A0L7REG0 A0A182R5F1 A0A0K8SEC3 A0A0K8SED0 A0A1B0CL85 A0A1J1HGP5 X1WIY0 A0A2S2NWN5 A0A2S2R5J2 A0A0P5C2J9 A0A0P5C2V6 A0A0P4XXH3 E9G466 A0A0P5J0H7 A0A0P5KKF5 A0A0P4XG62 A0A0P6JKJ8 A0A1J1HSY6 A0A182Y1T9 A0A3Q0IU93 A0A0P5CXW4 A0A2R5LHX7 A0A087UFI0 A0A293LPM2 V5GV24 A0A1B0B1B3 A0A0K2UVK0 B7QHM6 A0A0K2UW67 A0A023GAS5 A0A1E1XVG0 A0A1A9YEM1 A0A1V9XLZ5 A0A131Z3M2 A0A2P6L132 A0A0C9SEB1 T1JS33 T1J4X9
PDB
6H04
E-value=0.0462152,
Score=82
Ontologies
GO
PANTHER
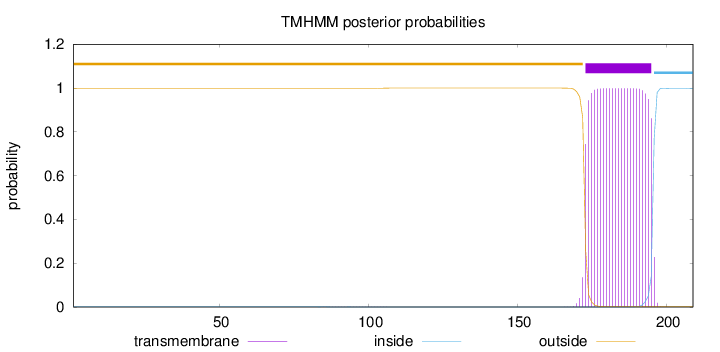
Topology
Subcellular location
Secreted
Length:
209
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.86226
Exp number, first 60 AAs:
0.00079
Total prob of N-in:
0.00087
outside
1 - 172
TMhelix
173 - 195
inside
196 - 209
Population Genetic Test Statistics
Pi
162.215543
Theta
155.655048
Tajima's D
0.233739
CLR
0.181981
CSRT
0.430278486075696
Interpretation
Uncertain
