Gene
KWMTBOMO01625
Pre Gene Modal
BGIBMGA006093
Annotation
PREDICTED:_kelch-like_protein_diablo_[Papilio_xuthus]
Full name
Kelch-like protein diablo
Location in the cell
Extracellular Reliability : 1.205 Lysosomal Reliability : 1.232
Sequence
CDS
ATGGGTGAGGGCGGATCCCCAGGTGGCGCGCGGCTGTCCCACACCTCGGAGAAACATCCGCGCGCCATACTCGGCGAACTCTCCGCACTGCGACGTCATCGTGAGCTATGCGATGTCGTGCTCAACGTTGCTAACAGGAAACTCTTTGCTCACAGAGTAATCCTATCAGCATGCAGCCCGTACTTCCGAGCGATGTTCACTGGGGAACTGGCCGAGTCTCGCGCCACCGAAGTCACGATCCGGGACGTGGACGAGCAGGCCATGGAGCAGCTCGTGGAGTTCTGCTACACCGCGCACATCGTCGTCGAGGAGAGTAACGTGCAGGCCCTGCTCCCGGCTGCGTGCCTGCTGCAGCTGCAAGAGATCCAGGACGTGTGCTGCGAGTTCCTCAAGCGTCAGCTGGACTGCTCCAACTGCCTCGGCATCAGAGCCTTCGCCGACACCCACTCGTGTCGGGAACTGCTCCGGATCGCCGATAAATTCACACAACAGAACTTCCCAGAGGTGATGGAGAGTGAAGAGTTCTTACTGCTGCCCGCCTCGCAGCTGATCGACATCGTGTCCTCGGACGAGCTGAACGTTCGCTCGGAGGAGCAGACTTTCCAGGCCGTCATGTCCTGGGTCAAGTACAACGTGGCCGAGCGGCGGCAGCACCTGGCGCAGGTGCTCCAGCACGTCCGACTGCCGCTGCTGAGTCCCAAGTTCCTGGTGGGCACCGTCTCCTCGGAGCTGCTGATCCGCTCCGACGACGCCTGCCGCGACCTGCTGGACGAGGCCAAGAACTACCTGCTGCTGCCGCAGGAGCGTCCGCTCATGCAGGGACCTAGGACTCGGCCCAGGAAGCCTACCAGGCGTGGCGAGGTGCTGTTCGCGGTGGGCGGCTGGTGCTCCGGGGACGCCATCGCGTCGGTGGAGCGGTTCGACCCGCAGAGCGGCGAGTGGAAGATGGTGGCGCCCATGTCCAAGCGGCGCTGCGGCGTGGGCGTGGCCGTGCTGCACGACCTGCTGTACGCAGTGGGCGGCCACGACGGACAGAGCTACCTCAACAGCATCGAGCGGTACGACCCGCAGACCAACCAGTGGTGCGGCGCCGTCGCGCCCACGTCGTCGTGCCGCACGTCCGTGGGCGTGGCCGTGCTGGAGGGCGCGCTGTACGCCGTGGGCGGGCAGGACGGCGTCCAGTGCCTCAACCACGTCGAGCGGTACGACCCCAAGGAGAACCGCTGGACCAAGGTCGCGCCCATGACCACGCGGCGTCTGGGCGTCGCCGTGGCAGTGCTGGGCGGACATCTGTACGCCGTGGGCGGCTCCGACGGGCAGGCGCCGCTCAACACCGTAGAGCGGTACGACCCGCGGCACAACAAGTGGACGCCCGTGGCCCCCATGTCCACCAGGAGGAAGCACCTGGGCTGCGCCGTGTTCGACGGGCAGATCTACGCCGTGGGGGGCCGGGACGACTGCACCGAGCTGTCCTCCGCCGAACGGTACGAGCCCAGCACGGACTCGTGGGCGCCCGTGGTGGCCATGACGTCGCGCCGGTCGGGCGTGGGGCTCGCCGTGGTCAACTCGCAGCTCTACGCCGTCGGGGGCTTCGACGGGACCGCCTACCTCAAGTCCATCGAGGTGTTCGATCCGGAATCGAACCAGTGGCGGCTGTGCGGCGCCATGAACTACCGGCGGCTCGGCGGCGGCGTCGGCGTCATGCGCTCGCCGCACCACGACACGCACTACATCTGGAACCGAAAAGATTCAGTGGTGTAA
Protein
MGEGGSPGGARLSHTSEKHPRAILGELSALRRHRELCDVVLNVANRKLFAHRVILSACSPYFRAMFTGELAESRATEVTIRDVDEQAMEQLVEFCYTAHIVVEESNVQALLPAACLLQLQEIQDVCCEFLKRQLDCSNCLGIRAFADTHSCRELLRIADKFTQQNFPEVMESEEFLLLPASQLIDIVSSDELNVRSEEQTFQAVMSWVKYNVAERRQHLAQVLQHVRLPLLSPKFLVGTVSSELLIRSDDACRDLLDEAKNYLLLPQERPLMQGPRTRPRKPTRRGEVLFAVGGWCSGDAIASVERFDPQSGEWKMVAPMSKRRCGVGVAVLHDLLYAVGGHDGQSYLNSIERYDPQTNQWCGAVAPTSSCRTSVGVAVLEGALYAVGGQDGVQCLNHVERYDPKENRWTKVAPMTTRRLGVAVAVLGGHLYAVGGSDGQAPLNTVERYDPRHNKWTPVAPMSTRRKHLGCAVFDGQIYAVGGRDDCTELSSAERYEPSTDSWAPVVAMTSRRSGVGLAVVNSQLYAVGGFDGTAYLKSIEVFDPESNQWRLCGAMNYRRLGGGVGVMRSPHHDTHYIWNRKDSVV
Summary
Description
Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. May have a role in synapse differentiation and growth (By similarity).
Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins (By similarity). May have a role in synapse differentiation and growth.
Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins (By similarity). May have a role in synapse differentiation and growth.
Keywords
Actin-binding
Complete proteome
Kelch repeat
Reference proteome
Repeat
Ubl conjugation pathway
Phosphoprotein
Feature
chain Kelch-like protein diablo
Uniprot
H9J9A1
A0A212F7Y2
A0A2A4K823
A0A2W1BX11
A0A2H1VFF1
W5J897
+ More
Q16RL8 A0A146KZV2 A0A1A9Y0X7 A0A1B0B6W7 A0A1A9W745 A0A2M4AEL0 A0A2M4BIR6 A0A0K8U4L8 A0A195F637 A0A158NLX1 A0A0A1WSQ3 M9PFL3 A0A1W4W686 A0A0C9RKS8 A0A1Y1KE61 A0A336LST5 A0A0C9R0D6 A0A0J9RW76 A0A182X7R8 A0A182MXQ1 A0A182WEQ6 A0A182HN73 A0A1L8DMN9 A0A195DB30 K7J3U8 A0A1B0CL92 A0A151X803 A0A1W4VB51 F4W735 E2A5N0 A0A0Q9XPU5 E9JBX9 A0A088AAP2 A0A1B0F0A0 A0A0N1IU24 A0A1I8Q6X6 A0A1I8N3N7 A0A154PMY4 A0A224X981 A0A0L7QR81 A0A067REC6 A0A182FBI1 A0A1W4WHX5 A0A1A9V3U5 A0A1B6D573 A0A182RAP5 A0A0N7Z9C2 A0A182UVU6 A0A1Q3G3J6 B0WWP2 W8BP65 A0A182PUL3 A0A1B0G8K6 A0A182LVF7 A0A195C1X6 A0A2M3ZHH3 A0A182UFW4 U5EMM9 A0A2J7RF21 A0A0R1DYG0 B4PD06 B4HIK1 Q9VUU5 A0A3B0KMK6 A0A2J7RF25 A0A0M4E9P4 A0A0Q5UK10 B3NDN0 B3M9V8 B4GRJ2 A0A0J9RVU5 B4QLQ2 A0A182IXM2 D6WNA5 A0A0R3P3S1 Q2M0J9 B4L0G9 B4J045 A0A0Q9WJY0 A0A1W4VMU2 B4LIG6 Q7QGL0 B4MXW3 A0A2A3EMJ6 A0A182Y623 A0A182QA14 A0A182LP13 A0A1S4H734 A0A182SAU3 A0A0P6E7W0 A0A3L8E441 A0A0P5ZBA5 E9FR39 A0A0P5DZD4
Q16RL8 A0A146KZV2 A0A1A9Y0X7 A0A1B0B6W7 A0A1A9W745 A0A2M4AEL0 A0A2M4BIR6 A0A0K8U4L8 A0A195F637 A0A158NLX1 A0A0A1WSQ3 M9PFL3 A0A1W4W686 A0A0C9RKS8 A0A1Y1KE61 A0A336LST5 A0A0C9R0D6 A0A0J9RW76 A0A182X7R8 A0A182MXQ1 A0A182WEQ6 A0A182HN73 A0A1L8DMN9 A0A195DB30 K7J3U8 A0A1B0CL92 A0A151X803 A0A1W4VB51 F4W735 E2A5N0 A0A0Q9XPU5 E9JBX9 A0A088AAP2 A0A1B0F0A0 A0A0N1IU24 A0A1I8Q6X6 A0A1I8N3N7 A0A154PMY4 A0A224X981 A0A0L7QR81 A0A067REC6 A0A182FBI1 A0A1W4WHX5 A0A1A9V3U5 A0A1B6D573 A0A182RAP5 A0A0N7Z9C2 A0A182UVU6 A0A1Q3G3J6 B0WWP2 W8BP65 A0A182PUL3 A0A1B0G8K6 A0A182LVF7 A0A195C1X6 A0A2M3ZHH3 A0A182UFW4 U5EMM9 A0A2J7RF21 A0A0R1DYG0 B4PD06 B4HIK1 Q9VUU5 A0A3B0KMK6 A0A2J7RF25 A0A0M4E9P4 A0A0Q5UK10 B3NDN0 B3M9V8 B4GRJ2 A0A0J9RVU5 B4QLQ2 A0A182IXM2 D6WNA5 A0A0R3P3S1 Q2M0J9 B4L0G9 B4J045 A0A0Q9WJY0 A0A1W4VMU2 B4LIG6 Q7QGL0 B4MXW3 A0A2A3EMJ6 A0A182Y623 A0A182QA14 A0A182LP13 A0A1S4H734 A0A182SAU3 A0A0P6E7W0 A0A3L8E441 A0A0P5ZBA5 E9FR39 A0A0P5DZD4
Pubmed
19121390
22118469
28756777
20920257
23761445
17510324
+ More
26823975 21347285 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 28004739 22936249 20075255 21719571 20798317 17994087 18057021 21282665 25315136 24845553 27129103 24495485 17550304 16408305 18327897 18362917 19820115 15632085 23185243 12364791 25244985 20966253 30249741 21292972
26823975 21347285 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 28004739 22936249 20075255 21719571 20798317 17994087 18057021 21282665 25315136 24845553 27129103 24495485 17550304 16408305 18327897 18362917 19820115 15632085 23185243 12364791 25244985 20966253 30249741 21292972
EMBL
BABH01004347
AGBW02009824
OWR49833.1
NWSH01000047
PCG80219.1
KZ149934
+ More
PZC77216.1 ODYU01002281 SOQ39555.1 ADMH02002103 ETN59095.1 CH477704 EAT37050.1 GDHC01016758 GDHC01002501 JAQ01871.1 JAQ16128.1 JXJN01009256 GGFK01005882 MBW39203.1 GGFJ01003567 MBW52708.1 GDHF01031636 GDHF01031089 GDHF01023387 GDHF01022982 GDHF01013229 GDHF01001251 JAI20678.1 JAI21225.1 JAI28927.1 JAI29332.1 JAI39085.1 JAI51063.1 KQ981805 KYN35637.1 ADTU01002193 GBXI01012752 JAD01540.1 AE014296 AGB94585.1 GBYB01007521 JAG77288.1 GEZM01091177 JAV57057.1 UFQS01000171 UFQT01000171 SSX00706.1 SSX21086.1 GBYB01006401 GBYB01006402 GBYB01006404 JAG76168.1 JAG76169.1 JAG76171.1 CM002912 KMY99832.1 APCN01001877 GFDF01006474 JAV07610.1 KQ981042 KYN10093.1 AJWK01017047 KQ982431 KYQ56493.1 GL887802 EGI70025.1 GL437023 EFN71227.1 CH933809 KRG06513.1 KRG06514.1 GL771642 EFZ09633.1 AJVK01007787 KQ435711 KOX79474.1 KQ434994 KZC13203.1 GFTR01007530 JAW08896.1 KQ414784 KOC61133.1 KK852514 KDR22201.1 GEDC01016446 GEDC01012954 JAS20852.1 JAS24344.1 GDKW01000955 JAI55640.1 GFDL01000667 JAV34378.1 DS232150 EDS36134.1 GAMC01008087 JAB98468.1 CCAG010000259 AXCM01004742 KQ978344 KYM94847.1 GGFM01007243 MBW27994.1 GANO01000954 JAB58917.1 NEVH01004414 PNF39435.1 CM000159 KRK02120.1 EDW94938.1 CH480815 EDW41631.1 AB027612 AF237711 BT010272 AAF49578.1 AAQ23590.1 BAD06413.1 OUUW01000009 SPP85038.1 PNF39436.1 CP012525 ALC43854.1 CH954178 KQS44130.1 EDV52163.1 CH902618 EDV40149.1 CH479188 EDW40377.1 KMY99833.1 CM000363 EDX10601.1 KQ971342 EFA03772.1 CH379069 KRT07769.1 KRT07770.1 EAL30933.2 EDW19138.1 CH916366 EDV97838.1 CH940647 KRF85085.1 EDW70753.1 AAAB01008834 CH963876 EDW76882.1 KZ288210 PBC32927.1 AXCN02001048 GDIQ01080991 JAN13746.1 QOIP01000001 RLU27185.1 GDIP01046201 JAM57514.1 GL732523 EFX90258.1 GDIP01150141 LRGB01000248 JAJ73261.1 KZS20043.1
PZC77216.1 ODYU01002281 SOQ39555.1 ADMH02002103 ETN59095.1 CH477704 EAT37050.1 GDHC01016758 GDHC01002501 JAQ01871.1 JAQ16128.1 JXJN01009256 GGFK01005882 MBW39203.1 GGFJ01003567 MBW52708.1 GDHF01031636 GDHF01031089 GDHF01023387 GDHF01022982 GDHF01013229 GDHF01001251 JAI20678.1 JAI21225.1 JAI28927.1 JAI29332.1 JAI39085.1 JAI51063.1 KQ981805 KYN35637.1 ADTU01002193 GBXI01012752 JAD01540.1 AE014296 AGB94585.1 GBYB01007521 JAG77288.1 GEZM01091177 JAV57057.1 UFQS01000171 UFQT01000171 SSX00706.1 SSX21086.1 GBYB01006401 GBYB01006402 GBYB01006404 JAG76168.1 JAG76169.1 JAG76171.1 CM002912 KMY99832.1 APCN01001877 GFDF01006474 JAV07610.1 KQ981042 KYN10093.1 AJWK01017047 KQ982431 KYQ56493.1 GL887802 EGI70025.1 GL437023 EFN71227.1 CH933809 KRG06513.1 KRG06514.1 GL771642 EFZ09633.1 AJVK01007787 KQ435711 KOX79474.1 KQ434994 KZC13203.1 GFTR01007530 JAW08896.1 KQ414784 KOC61133.1 KK852514 KDR22201.1 GEDC01016446 GEDC01012954 JAS20852.1 JAS24344.1 GDKW01000955 JAI55640.1 GFDL01000667 JAV34378.1 DS232150 EDS36134.1 GAMC01008087 JAB98468.1 CCAG010000259 AXCM01004742 KQ978344 KYM94847.1 GGFM01007243 MBW27994.1 GANO01000954 JAB58917.1 NEVH01004414 PNF39435.1 CM000159 KRK02120.1 EDW94938.1 CH480815 EDW41631.1 AB027612 AF237711 BT010272 AAF49578.1 AAQ23590.1 BAD06413.1 OUUW01000009 SPP85038.1 PNF39436.1 CP012525 ALC43854.1 CH954178 KQS44130.1 EDV52163.1 CH902618 EDV40149.1 CH479188 EDW40377.1 KMY99833.1 CM000363 EDX10601.1 KQ971342 EFA03772.1 CH379069 KRT07769.1 KRT07770.1 EAL30933.2 EDW19138.1 CH916366 EDV97838.1 CH940647 KRF85085.1 EDW70753.1 AAAB01008834 CH963876 EDW76882.1 KZ288210 PBC32927.1 AXCN02001048 GDIQ01080991 JAN13746.1 QOIP01000001 RLU27185.1 GDIP01046201 JAM57514.1 GL732523 EFX90258.1 GDIP01150141 LRGB01000248 JAJ73261.1 KZS20043.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000000673
UP000008820
UP000092443
+ More
UP000092460 UP000091820 UP000078541 UP000005205 UP000000803 UP000192223 UP000076407 UP000075884 UP000075920 UP000075840 UP000078492 UP000002358 UP000092461 UP000075809 UP000192221 UP000007755 UP000000311 UP000009192 UP000005203 UP000092462 UP000053105 UP000095300 UP000095301 UP000076502 UP000053825 UP000027135 UP000069272 UP000078200 UP000075900 UP000075903 UP000002320 UP000075885 UP000092444 UP000075883 UP000078542 UP000075902 UP000235965 UP000002282 UP000001292 UP000268350 UP000092553 UP000008711 UP000007801 UP000008744 UP000000304 UP000075880 UP000007266 UP000001819 UP000001070 UP000008792 UP000007062 UP000007798 UP000242457 UP000076408 UP000075886 UP000075882 UP000075901 UP000279307 UP000000305 UP000076858
UP000092460 UP000091820 UP000078541 UP000005205 UP000000803 UP000192223 UP000076407 UP000075884 UP000075920 UP000075840 UP000078492 UP000002358 UP000092461 UP000075809 UP000192221 UP000007755 UP000000311 UP000009192 UP000005203 UP000092462 UP000053105 UP000095300 UP000095301 UP000076502 UP000053825 UP000027135 UP000069272 UP000078200 UP000075900 UP000075903 UP000002320 UP000075885 UP000092444 UP000075883 UP000078542 UP000075902 UP000235965 UP000002282 UP000001292 UP000268350 UP000092553 UP000008711 UP000007801 UP000008744 UP000000304 UP000075880 UP000007266 UP000001819 UP000001070 UP000008792 UP000007062 UP000007798 UP000242457 UP000076408 UP000075886 UP000075882 UP000075901 UP000279307 UP000000305 UP000076858
Interpro
Gene 3D
ProteinModelPortal
H9J9A1
A0A212F7Y2
A0A2A4K823
A0A2W1BX11
A0A2H1VFF1
W5J897
+ More
Q16RL8 A0A146KZV2 A0A1A9Y0X7 A0A1B0B6W7 A0A1A9W745 A0A2M4AEL0 A0A2M4BIR6 A0A0K8U4L8 A0A195F637 A0A158NLX1 A0A0A1WSQ3 M9PFL3 A0A1W4W686 A0A0C9RKS8 A0A1Y1KE61 A0A336LST5 A0A0C9R0D6 A0A0J9RW76 A0A182X7R8 A0A182MXQ1 A0A182WEQ6 A0A182HN73 A0A1L8DMN9 A0A195DB30 K7J3U8 A0A1B0CL92 A0A151X803 A0A1W4VB51 F4W735 E2A5N0 A0A0Q9XPU5 E9JBX9 A0A088AAP2 A0A1B0F0A0 A0A0N1IU24 A0A1I8Q6X6 A0A1I8N3N7 A0A154PMY4 A0A224X981 A0A0L7QR81 A0A067REC6 A0A182FBI1 A0A1W4WHX5 A0A1A9V3U5 A0A1B6D573 A0A182RAP5 A0A0N7Z9C2 A0A182UVU6 A0A1Q3G3J6 B0WWP2 W8BP65 A0A182PUL3 A0A1B0G8K6 A0A182LVF7 A0A195C1X6 A0A2M3ZHH3 A0A182UFW4 U5EMM9 A0A2J7RF21 A0A0R1DYG0 B4PD06 B4HIK1 Q9VUU5 A0A3B0KMK6 A0A2J7RF25 A0A0M4E9P4 A0A0Q5UK10 B3NDN0 B3M9V8 B4GRJ2 A0A0J9RVU5 B4QLQ2 A0A182IXM2 D6WNA5 A0A0R3P3S1 Q2M0J9 B4L0G9 B4J045 A0A0Q9WJY0 A0A1W4VMU2 B4LIG6 Q7QGL0 B4MXW3 A0A2A3EMJ6 A0A182Y623 A0A182QA14 A0A182LP13 A0A1S4H734 A0A182SAU3 A0A0P6E7W0 A0A3L8E441 A0A0P5ZBA5 E9FR39 A0A0P5DZD4
Q16RL8 A0A146KZV2 A0A1A9Y0X7 A0A1B0B6W7 A0A1A9W745 A0A2M4AEL0 A0A2M4BIR6 A0A0K8U4L8 A0A195F637 A0A158NLX1 A0A0A1WSQ3 M9PFL3 A0A1W4W686 A0A0C9RKS8 A0A1Y1KE61 A0A336LST5 A0A0C9R0D6 A0A0J9RW76 A0A182X7R8 A0A182MXQ1 A0A182WEQ6 A0A182HN73 A0A1L8DMN9 A0A195DB30 K7J3U8 A0A1B0CL92 A0A151X803 A0A1W4VB51 F4W735 E2A5N0 A0A0Q9XPU5 E9JBX9 A0A088AAP2 A0A1B0F0A0 A0A0N1IU24 A0A1I8Q6X6 A0A1I8N3N7 A0A154PMY4 A0A224X981 A0A0L7QR81 A0A067REC6 A0A182FBI1 A0A1W4WHX5 A0A1A9V3U5 A0A1B6D573 A0A182RAP5 A0A0N7Z9C2 A0A182UVU6 A0A1Q3G3J6 B0WWP2 W8BP65 A0A182PUL3 A0A1B0G8K6 A0A182LVF7 A0A195C1X6 A0A2M3ZHH3 A0A182UFW4 U5EMM9 A0A2J7RF21 A0A0R1DYG0 B4PD06 B4HIK1 Q9VUU5 A0A3B0KMK6 A0A2J7RF25 A0A0M4E9P4 A0A0Q5UK10 B3NDN0 B3M9V8 B4GRJ2 A0A0J9RVU5 B4QLQ2 A0A182IXM2 D6WNA5 A0A0R3P3S1 Q2M0J9 B4L0G9 B4J045 A0A0Q9WJY0 A0A1W4VMU2 B4LIG6 Q7QGL0 B4MXW3 A0A2A3EMJ6 A0A182Y623 A0A182QA14 A0A182LP13 A0A1S4H734 A0A182SAU3 A0A0P6E7W0 A0A3L8E441 A0A0P5ZBA5 E9FR39 A0A0P5DZD4
PDB
6GY5
E-value=1.01178e-139,
Score=1275
Ontologies
GO
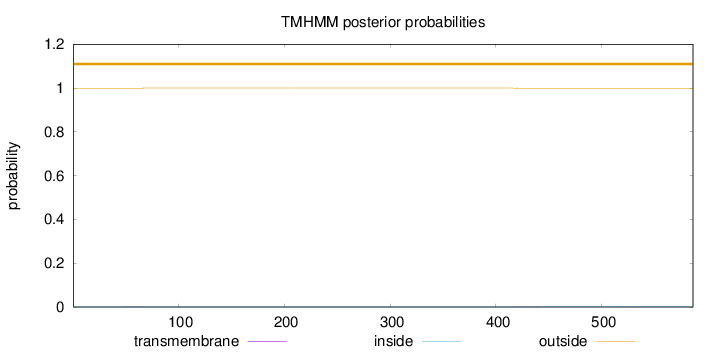
Topology
Length:
586
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02621
Exp number, first 60 AAs:
0.0038
Total prob of N-in:
0.00048
outside
1 - 586
Population Genetic Test Statistics
Pi
52.740922
Theta
176.531162
Tajima's D
-1.497267
CLR
1.602417
CSRT
0.063896805159742
Interpretation
Possibly Positive selection
