Pre Gene Modal
BGIBMGA006072
Annotation
PREDICTED:_conserved_oligomeric_Golgi_complex_subunit_4-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.281
Sequence
CDS
ATGTCGCTTAGTTTATTACTTGAGAAATATGATGTATCAACAGAAGAGGGCTTACAAAAAGCGTTAAGCGAAGTTGAAAAAGAAGAATCAGAAGTAGATGAAGCATTATCTGGAGTGCTATCGCGTGCTTGCATGCTAGAAGACAAGCTCCGCACAGCCAGTCAAGCTTACACTAAGCTCAGTGAAGTGAAAGCTGATGCACAAGTTGCTGCAGACATGGTGGACAGAACTGCAGAATTAGCCAGAGATGTCAGTGCTAAAGTCAGACAACTCGATTTGGCTAGATCTAGAGTGGCAGAATGTCAGAGGAGAGTGCATGATCTTATTGATCTGCAGCTATGCAGCGCAGGAGTTGAAGCTGCCATAAAAGCTAATGATTATGAGACCGGTGCTGGACACGTGTCGAGATTCCTAAGCATGGATCCAGGTTCAGTAGCGGCAGCGCGGGCTCGCGGAGCCGCCGACCCACGGGACGCAATGACGCGCGCCGCCAACACACTCAGAGAACATATCGTTCGTAGGTTTGAAGATGCCGCTCAGAAAAACGATGACGAGAACGTAGATAAGCTTTTCAAGCTGTTTCCTCAGATCGGACGCGCCGAGGAAGGAGTCCGGCTGCTGGCCAAGTACCTGGCCACCAAGTTGGGGGTGGAGATCCGTCGGCTGAGCGTGGTGGCGGAGGGCGAGGGCGCCGTGTTCGCCGACGCCATGACGCGCGTGCTGGAGGCCGCCGCCGCCGCCGCCGCGCGCGCCGCCGCCCTCGCCGCCCTCGCCGCGCCCGGCCTGCTGCCCGCCGCGCTCGCCGCCCTGCAGCCGCAGGTCCGTCTCACCTTCACTTCATTATATACACTTTTTATGGTGAAGGGAAAAAATCAAGAAATCGCATGA
Protein
MSLSLLLEKYDVSTEEGLQKALSEVEKEESEVDEALSGVLSRACMLEDKLRTASQAYTKLSEVKADAQVAADMVDRTAELARDVSAKVRQLDLARSRVAECQRRVHDLIDLQLCSAGVEAAIKANDYETGAGHVSRFLSMDPGSVAAARARGAADPRDAMTRAANTLREHIVRRFEDAAQKNDDENVDKLFKLFPQIGRAEEGVRLLAKYLATKLGVEIRRLSVVAEGEGAVFADAMTRVLEAAAAAAARAAALAALAAPGLLPAALAALQPQVRLTFTSLYTLFMVKGKNQEIA
Summary
Uniprot
H9J980
A0A2W1BVJ5
A0A2A4K8J4
A0A2H1VH18
S4PSG0
A0A194Q0D8
+ More
A0A194RBL9 A0A212F7W5 U5EZ33 A0A182JSD8 A0A1S4GZG6 A0A182U1B3 Q5TNE6 A0A182VEV9 A0A182RUR1 A0A182SI44 A0A182XDD6 A0A182LMZ7 A0A182H6I8 A0A182VU97 A0A182P685 A0A182Y942 A0A1B0C0R4 A0A1B0FCV5 A0A182M436 A0A1A9YH38 Q16TS8 A0A1A9VI52 A0A1A9ZEX6 A0A182I766 A0A1A9WLY3 B0WBZ5 A0A084VYM5 B4LVB3 A0A182F5R5 A0A2M4APT9 A0A1I8P8E2 A0A1B6E410 B4JQ08 A0A336KIL1 A0A088A322 A0A182IKX9 A0A067R0K6 A0A182QER7 W5J1S1 B4KHQ6 A0A1B6DE98 A0A154P3H1 A0A0C9PZX8 A0A195EFP0 A0A0L7QXE7 A0A151WJK3 E9IBA8 A0A2J7PPJ7 A0A0M9A9M2 A0A2J7PPG1 A0A310SQB6 A0A2J7PPI9 A0A158NI43 A0A195B651 A0A026WSF9 A0A1L8DVE8 A0A182NGK8 A0A0J7P510 F4WGD4 E2BV79 A0A195EZM1 A0A1B6GDQ7 W8CBU5 A0A0R3NW10 A0A0M5J668 A0A1B0GIM2 A0A3B0JHN3 A0A195CP06 A0A1B6LM77 A0A1J1I1I0 A0A1I8MWB7 B4MUA5 B3MJJ3 A0A0P5YEW4 A0A0P6EHI8 A0A0P5BBP6 A0A0P5B8U0 A0A0P4Y3D0 E9FQQ8 A0A0P5LU46 A0A0N7ZH79 A0A0P6I8Q9 A0A0P5RJU6 A0A0P5DTD5 A0A0K8V4A8 A0A1W4UT09 A0A0A1WXE4
A0A194RBL9 A0A212F7W5 U5EZ33 A0A182JSD8 A0A1S4GZG6 A0A182U1B3 Q5TNE6 A0A182VEV9 A0A182RUR1 A0A182SI44 A0A182XDD6 A0A182LMZ7 A0A182H6I8 A0A182VU97 A0A182P685 A0A182Y942 A0A1B0C0R4 A0A1B0FCV5 A0A182M436 A0A1A9YH38 Q16TS8 A0A1A9VI52 A0A1A9ZEX6 A0A182I766 A0A1A9WLY3 B0WBZ5 A0A084VYM5 B4LVB3 A0A182F5R5 A0A2M4APT9 A0A1I8P8E2 A0A1B6E410 B4JQ08 A0A336KIL1 A0A088A322 A0A182IKX9 A0A067R0K6 A0A182QER7 W5J1S1 B4KHQ6 A0A1B6DE98 A0A154P3H1 A0A0C9PZX8 A0A195EFP0 A0A0L7QXE7 A0A151WJK3 E9IBA8 A0A2J7PPJ7 A0A0M9A9M2 A0A2J7PPG1 A0A310SQB6 A0A2J7PPI9 A0A158NI43 A0A195B651 A0A026WSF9 A0A1L8DVE8 A0A182NGK8 A0A0J7P510 F4WGD4 E2BV79 A0A195EZM1 A0A1B6GDQ7 W8CBU5 A0A0R3NW10 A0A0M5J668 A0A1B0GIM2 A0A3B0JHN3 A0A195CP06 A0A1B6LM77 A0A1J1I1I0 A0A1I8MWB7 B4MUA5 B3MJJ3 A0A0P5YEW4 A0A0P6EHI8 A0A0P5BBP6 A0A0P5B8U0 A0A0P4Y3D0 E9FQQ8 A0A0P5LU46 A0A0N7ZH79 A0A0P6I8Q9 A0A0P5RJU6 A0A0P5DTD5 A0A0K8V4A8 A0A1W4UT09 A0A0A1WXE4
Pubmed
EMBL
BABH01004345
BABH01004346
KZ149934
PZC77217.1
NWSH01000047
PCG80238.1
+ More
ODYU01002281 SOQ39554.1 GAIX01013853 JAA78707.1 KQ459584 KPI98459.1 KQ460644 KPJ13296.1 AGBW02009824 OWR49834.1 GANO01001515 JAB58356.1 AAAB01008984 EAL38955.3 JXUM01114158 KQ565759 KXJ70693.1 JXJN01023734 CCAG010017495 AXCM01000336 CH477641 EAT37914.1 APCN01003368 DS231884 EDS43055.1 ATLV01018379 KE525231 KFB43069.1 CH940649 EDW63292.2 GGFK01009484 MBW42805.1 GEDC01004650 JAS32648.1 CH916372 EDV98988.1 UFQS01000186 UFQT01000186 SSX00900.1 SSX21280.1 KK853129 KDR10987.1 AXCN02002086 ADMH02002160 ETN58187.1 CH933807 EDW13343.2 GEDC01013270 JAS24028.1 KQ434809 KZC06397.1 GBYB01007118 GBYB01011682 JAG76885.1 JAG81449.1 KQ978957 KYN27090.1 KQ414704 KOC63292.1 KQ983039 KYQ48011.1 GL762111 EFZ22111.1 NEVH01022696 PNF18229.1 KQ435719 KOX78763.1 PNF18230.1 KQ761039 OAD58347.1 PNF18231.1 ADTU01016268 KQ976579 KYM79993.1 KK107111 QOIP01000011 EZA58967.1 RLU17155.1 GFDF01003684 JAV10400.1 LBMM01000092 KMR04999.1 GL888128 EGI66901.1 GL450824 EFN80371.1 KQ981897 KYN33750.1 GECZ01009212 JAS60557.1 GAMC01004511 JAC02045.1 CH475452 KRT05329.1 CP012523 ALC39286.1 AJWK01015079 AJWK01015080 OUUW01000006 SPP81705.1 KQ977481 KYN02446.1 GEBQ01015127 GEBQ01002337 JAT24850.1 JAT37640.1 CVRI01000038 CRK94184.1 CH963857 EDW76031.1 CH902620 EDV32361.1 GDIP01072795 GDIQ01037812 JAM30920.1 JAN56925.1 GDIQ01069347 JAN25390.1 GDIP01190986 JAJ32416.1 GDIP01188434 JAJ34968.1 GDIP01233272 JAI90129.1 GL732523 EFX90030.1 GDIQ01165560 JAK86165.1 GDIP01245705 JAI77696.1 GDIQ01009669 JAN85068.1 GDIQ01101482 JAL50244.1 GDIP01151535 JAJ71867.1 GDHF01018894 JAI33420.1 GBXI01011199 JAD03093.1
ODYU01002281 SOQ39554.1 GAIX01013853 JAA78707.1 KQ459584 KPI98459.1 KQ460644 KPJ13296.1 AGBW02009824 OWR49834.1 GANO01001515 JAB58356.1 AAAB01008984 EAL38955.3 JXUM01114158 KQ565759 KXJ70693.1 JXJN01023734 CCAG010017495 AXCM01000336 CH477641 EAT37914.1 APCN01003368 DS231884 EDS43055.1 ATLV01018379 KE525231 KFB43069.1 CH940649 EDW63292.2 GGFK01009484 MBW42805.1 GEDC01004650 JAS32648.1 CH916372 EDV98988.1 UFQS01000186 UFQT01000186 SSX00900.1 SSX21280.1 KK853129 KDR10987.1 AXCN02002086 ADMH02002160 ETN58187.1 CH933807 EDW13343.2 GEDC01013270 JAS24028.1 KQ434809 KZC06397.1 GBYB01007118 GBYB01011682 JAG76885.1 JAG81449.1 KQ978957 KYN27090.1 KQ414704 KOC63292.1 KQ983039 KYQ48011.1 GL762111 EFZ22111.1 NEVH01022696 PNF18229.1 KQ435719 KOX78763.1 PNF18230.1 KQ761039 OAD58347.1 PNF18231.1 ADTU01016268 KQ976579 KYM79993.1 KK107111 QOIP01000011 EZA58967.1 RLU17155.1 GFDF01003684 JAV10400.1 LBMM01000092 KMR04999.1 GL888128 EGI66901.1 GL450824 EFN80371.1 KQ981897 KYN33750.1 GECZ01009212 JAS60557.1 GAMC01004511 JAC02045.1 CH475452 KRT05329.1 CP012523 ALC39286.1 AJWK01015079 AJWK01015080 OUUW01000006 SPP81705.1 KQ977481 KYN02446.1 GEBQ01015127 GEBQ01002337 JAT24850.1 JAT37640.1 CVRI01000038 CRK94184.1 CH963857 EDW76031.1 CH902620 EDV32361.1 GDIP01072795 GDIQ01037812 JAM30920.1 JAN56925.1 GDIQ01069347 JAN25390.1 GDIP01190986 JAJ32416.1 GDIP01188434 JAJ34968.1 GDIP01233272 JAI90129.1 GL732523 EFX90030.1 GDIQ01165560 JAK86165.1 GDIP01245705 JAI77696.1 GDIQ01009669 JAN85068.1 GDIQ01101482 JAL50244.1 GDIP01151535 JAJ71867.1 GDHF01018894 JAI33420.1 GBXI01011199 JAD03093.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000075881
+ More
UP000007062 UP000075902 UP000075903 UP000075900 UP000075901 UP000076407 UP000075882 UP000069940 UP000249989 UP000075920 UP000075885 UP000076408 UP000092460 UP000092444 UP000075883 UP000092443 UP000008820 UP000078200 UP000092445 UP000075840 UP000091820 UP000002320 UP000030765 UP000008792 UP000069272 UP000095300 UP000001070 UP000005203 UP000075880 UP000027135 UP000075886 UP000000673 UP000009192 UP000076502 UP000078492 UP000053825 UP000075809 UP000235965 UP000053105 UP000005205 UP000078540 UP000053097 UP000279307 UP000075884 UP000036403 UP000007755 UP000008237 UP000078541 UP000001819 UP000092553 UP000092461 UP000268350 UP000078542 UP000183832 UP000095301 UP000007798 UP000007801 UP000000305 UP000192221
UP000007062 UP000075902 UP000075903 UP000075900 UP000075901 UP000076407 UP000075882 UP000069940 UP000249989 UP000075920 UP000075885 UP000076408 UP000092460 UP000092444 UP000075883 UP000092443 UP000008820 UP000078200 UP000092445 UP000075840 UP000091820 UP000002320 UP000030765 UP000008792 UP000069272 UP000095300 UP000001070 UP000005203 UP000075880 UP000027135 UP000075886 UP000000673 UP000009192 UP000076502 UP000078492 UP000053825 UP000075809 UP000235965 UP000053105 UP000005205 UP000078540 UP000053097 UP000279307 UP000075884 UP000036403 UP000007755 UP000008237 UP000078541 UP000001819 UP000092553 UP000092461 UP000268350 UP000078542 UP000183832 UP000095301 UP000007798 UP000007801 UP000000305 UP000192221
PRIDE
Interpro
ProteinModelPortal
H9J980
A0A2W1BVJ5
A0A2A4K8J4
A0A2H1VH18
S4PSG0
A0A194Q0D8
+ More
A0A194RBL9 A0A212F7W5 U5EZ33 A0A182JSD8 A0A1S4GZG6 A0A182U1B3 Q5TNE6 A0A182VEV9 A0A182RUR1 A0A182SI44 A0A182XDD6 A0A182LMZ7 A0A182H6I8 A0A182VU97 A0A182P685 A0A182Y942 A0A1B0C0R4 A0A1B0FCV5 A0A182M436 A0A1A9YH38 Q16TS8 A0A1A9VI52 A0A1A9ZEX6 A0A182I766 A0A1A9WLY3 B0WBZ5 A0A084VYM5 B4LVB3 A0A182F5R5 A0A2M4APT9 A0A1I8P8E2 A0A1B6E410 B4JQ08 A0A336KIL1 A0A088A322 A0A182IKX9 A0A067R0K6 A0A182QER7 W5J1S1 B4KHQ6 A0A1B6DE98 A0A154P3H1 A0A0C9PZX8 A0A195EFP0 A0A0L7QXE7 A0A151WJK3 E9IBA8 A0A2J7PPJ7 A0A0M9A9M2 A0A2J7PPG1 A0A310SQB6 A0A2J7PPI9 A0A158NI43 A0A195B651 A0A026WSF9 A0A1L8DVE8 A0A182NGK8 A0A0J7P510 F4WGD4 E2BV79 A0A195EZM1 A0A1B6GDQ7 W8CBU5 A0A0R3NW10 A0A0M5J668 A0A1B0GIM2 A0A3B0JHN3 A0A195CP06 A0A1B6LM77 A0A1J1I1I0 A0A1I8MWB7 B4MUA5 B3MJJ3 A0A0P5YEW4 A0A0P6EHI8 A0A0P5BBP6 A0A0P5B8U0 A0A0P4Y3D0 E9FQQ8 A0A0P5LU46 A0A0N7ZH79 A0A0P6I8Q9 A0A0P5RJU6 A0A0P5DTD5 A0A0K8V4A8 A0A1W4UT09 A0A0A1WXE4
A0A194RBL9 A0A212F7W5 U5EZ33 A0A182JSD8 A0A1S4GZG6 A0A182U1B3 Q5TNE6 A0A182VEV9 A0A182RUR1 A0A182SI44 A0A182XDD6 A0A182LMZ7 A0A182H6I8 A0A182VU97 A0A182P685 A0A182Y942 A0A1B0C0R4 A0A1B0FCV5 A0A182M436 A0A1A9YH38 Q16TS8 A0A1A9VI52 A0A1A9ZEX6 A0A182I766 A0A1A9WLY3 B0WBZ5 A0A084VYM5 B4LVB3 A0A182F5R5 A0A2M4APT9 A0A1I8P8E2 A0A1B6E410 B4JQ08 A0A336KIL1 A0A088A322 A0A182IKX9 A0A067R0K6 A0A182QER7 W5J1S1 B4KHQ6 A0A1B6DE98 A0A154P3H1 A0A0C9PZX8 A0A195EFP0 A0A0L7QXE7 A0A151WJK3 E9IBA8 A0A2J7PPJ7 A0A0M9A9M2 A0A2J7PPG1 A0A310SQB6 A0A2J7PPI9 A0A158NI43 A0A195B651 A0A026WSF9 A0A1L8DVE8 A0A182NGK8 A0A0J7P510 F4WGD4 E2BV79 A0A195EZM1 A0A1B6GDQ7 W8CBU5 A0A0R3NW10 A0A0M5J668 A0A1B0GIM2 A0A3B0JHN3 A0A195CP06 A0A1B6LM77 A0A1J1I1I0 A0A1I8MWB7 B4MUA5 B3MJJ3 A0A0P5YEW4 A0A0P6EHI8 A0A0P5BBP6 A0A0P5B8U0 A0A0P4Y3D0 E9FQQ8 A0A0P5LU46 A0A0N7ZH79 A0A0P6I8Q9 A0A0P5RJU6 A0A0P5DTD5 A0A0K8V4A8 A0A1W4UT09 A0A0A1WXE4
Ontologies
GO
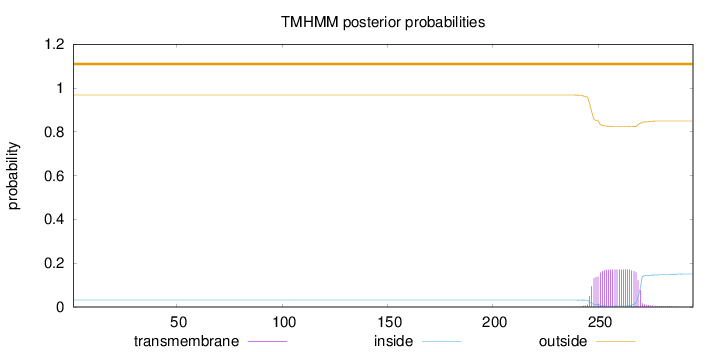
Topology
Length:
295
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.90174
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03197
outside
1 - 295
Population Genetic Test Statistics
Pi
145.652479
Theta
169.572354
Tajima's D
-0.807505
CLR
0.208409
CSRT
0.173341332933353
Interpretation
Possibly Positive selection
