Pre Gene Modal
BGIBMGA006091
Annotation
PREDICTED:_uncharacterized_protein_LOC106140453_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.999
Sequence
CDS
ATGTTTGATAATTTCCACAACAATCAGACTGTGGCACAAGAGCCCCCACCTCCTAAGAAAGTGCTTAGATGTGACAAGAGCTATGTCACAACTATACCAGGAATCCTGAAAGTTGTTCAAGTGTGCTGCAACATAATAGGCTTCATCTGTATCAAAGTGAGCTGGTCATGGGTATCGGCAATATTCTACAACATGGTATACTGGATTGGAAACATTCTGACTATTCTACTTCTTCTCGCATACACTTGTCATTTTGTTGAGAAGTATGACCACTGGCCTTGGTTCAAATATGAGTTCTTCTACTGCAGTATTATGGCCTTAGCTTATGTTGTGTGTTCAATATTTGCTGCTACCATTGGGGAAAGTGTTGGATATGCTGTTGTTTTCTTTGGATTGGTGGCAACCTGTGCCTATGGATTTGATGCTTACCAAAAATACAAAGGATGGAACAGGGGTCTGCCACCACAATGA
Protein
MFDNFHNNQTVAQEPPPPKKVLRCDKSYVTTIPGILKVVQVCCNIIGFICIKVSWSWVSAIFYNMVYWIGNILTILLLLAYTCHFVEKYDHWPWFKYEFFYCSIMALAYVVCSIFAATIGESVGYAVVFFGLVATCAYGFDAYQKYKGWNRGLPPQ
Summary
Uniprot
H9J999
A0A1E1W4N4
A0A2W1BSB3
A0A2H1WNT9
A0A2A4K8K1
A0A212F7Y8
+ More
A0A194PZI8 A0A0L7KMT7 A0A194R7T3 A0A212F7X2 A0A1W4W5Z0 A0A194Q4W7 A0A2W1BQ69 A0A195BX80 A0A158NCE3 A0A151WMV4 A0A2A4K8A6 F4WWY3 A0A151JQZ3 A0A195F9K1 A0A194R702 A0A182F5D6 A0A0K8TT38 A0A194Q0D3 A0A1E1WBQ1 A0A1E1W281 E2BBX3 A0A2A3E8E4 V9IB08 A0A182M1Q6 E2AYQ6 A0A154PNY3 W5JLN5 A0A2M3ZIN6 A0A2M4AV21 A0A182JF51 T1E3V1 A0A026VX22 A0A0T6AZD7 A0A336M2L0 A0A084WBY4 A0A087ZV46 A0A182T2H6 A0A0L7RG42 A0A182R9L1 Q16YL7 A0A023EI88 A0A182GVL0 A0A182YFB6 A0A182NZW2 A0A1Q3FFQ2 N6UFL6 A0A182I524 A0A182V9P4 A0A182WYP8 Q7PW02 A0A1B0C907 A0A0C9RNK5 T1JEB9 A0A182UCD8 A0A131XIV8 A0A1B6CLX8 A0A1L8E2T8 A0A182WH26 A0A2A4K7I5 A0A1J1IK14 A0A2W1BWN8 A0A1B0D093 U5EQI3 A0A2H1VFC6 G3ML46 A0A023G7D4 A0A0J7K9U5 A0A1B6KCF7 A0A151IBZ1 A0A1E1WZ53 D6WJ80 V5IGH3 A0A2J7QBU4 A0A087UEM3 A0A0A9W091 A0A1B6IUY1 A0A194R652 A0A232F6D9 I4DNI5 L7M764 A0A162QYT6 T1KTW6 A0A224YLQ2 A0A131YTR3 A0A131YI05 K7IZL1 A0A0P6IQR9 A0A2H1W4X8 A0A1B6EXG7 T1E2H4 A0A2R5L6S4 B0W1H7 A0A023F1B3
A0A194PZI8 A0A0L7KMT7 A0A194R7T3 A0A212F7X2 A0A1W4W5Z0 A0A194Q4W7 A0A2W1BQ69 A0A195BX80 A0A158NCE3 A0A151WMV4 A0A2A4K8A6 F4WWY3 A0A151JQZ3 A0A195F9K1 A0A194R702 A0A182F5D6 A0A0K8TT38 A0A194Q0D3 A0A1E1WBQ1 A0A1E1W281 E2BBX3 A0A2A3E8E4 V9IB08 A0A182M1Q6 E2AYQ6 A0A154PNY3 W5JLN5 A0A2M3ZIN6 A0A2M4AV21 A0A182JF51 T1E3V1 A0A026VX22 A0A0T6AZD7 A0A336M2L0 A0A084WBY4 A0A087ZV46 A0A182T2H6 A0A0L7RG42 A0A182R9L1 Q16YL7 A0A023EI88 A0A182GVL0 A0A182YFB6 A0A182NZW2 A0A1Q3FFQ2 N6UFL6 A0A182I524 A0A182V9P4 A0A182WYP8 Q7PW02 A0A1B0C907 A0A0C9RNK5 T1JEB9 A0A182UCD8 A0A131XIV8 A0A1B6CLX8 A0A1L8E2T8 A0A182WH26 A0A2A4K7I5 A0A1J1IK14 A0A2W1BWN8 A0A1B0D093 U5EQI3 A0A2H1VFC6 G3ML46 A0A023G7D4 A0A0J7K9U5 A0A1B6KCF7 A0A151IBZ1 A0A1E1WZ53 D6WJ80 V5IGH3 A0A2J7QBU4 A0A087UEM3 A0A0A9W091 A0A1B6IUY1 A0A194R652 A0A232F6D9 I4DNI5 L7M764 A0A162QYT6 T1KTW6 A0A224YLQ2 A0A131YTR3 A0A131YI05 K7IZL1 A0A0P6IQR9 A0A2H1W4X8 A0A1B6EXG7 T1E2H4 A0A2R5L6S4 B0W1H7 A0A023F1B3
Pubmed
19121390
28756777
22118469
26354079
26227816
21347285
+ More
21719571 26369729 20798317 20920257 23761445 24330624 24508170 30249741 24438588 17510324 24945155 26483478 25244985 23537049 12364791 14747013 17210077 28049606 22216098 28503490 18362917 19820115 25765539 25401762 26823975 28648823 22651552 25576852 28797301 26830274 20075255 25474469
21719571 26369729 20798317 20920257 23761445 24330624 24508170 30249741 24438588 17510324 24945155 26483478 25244985 23537049 12364791 14747013 17210077 28049606 22216098 28503490 18362917 19820115 25765539 25401762 26823975 28648823 22651552 25576852 28797301 26830274 20075255 25474469
EMBL
BABH01004342
GDQN01009118
JAT81936.1
KZ149934
PZC77221.1
ODYU01009945
+ More
SOQ54698.1 NWSH01000047 PCG80234.1 AGBW02009824 OWR49838.1 KQ459584 KPI98453.1 JTDY01008744 KOB64425.1 KQ460644 KPJ13300.1 OWR49837.1 KPI98455.1 PZC77219.1 KQ976396 KYM92900.1 ADTU01011823 KQ982934 KYQ49150.1 PCG80236.1 GL888417 EGI61226.1 KQ978619 KYN29786.1 KQ981727 KYN37056.1 KPJ13299.1 GDAI01000074 JAI17529.1 KPI98454.1 GDQN01006658 JAT84396.1 GDQN01010932 GDQN01010586 GDQN01009981 GDQN01006844 GDQN01006638 GDQN01004581 GDQN01002001 JAT80122.1 JAT80468.1 JAT81073.1 JAT84210.1 JAT84416.1 JAT86473.1 JAT89053.1 GL447175 EFN86797.1 KZ288348 PBC27476.1 JR038487 AEY58275.1 AXCM01006387 GL443983 EFN61419.1 KQ435007 KZC13591.1 ADMH02000797 ETN65036.1 GGFM01007584 MBW28335.1 GGFK01011318 MBW44639.1 GALA01000040 JAA94812.1 KK107648 QOIP01000008 EZA48307.1 RLU19681.1 LJIG01022548 KRT79991.1 UFQT01000438 SSX24310.1 ATLV01022519 KE525333 KFB47728.1 KQ414598 KOC69815.1 CH477514 EAT39718.1 GAPW01004972 JAC08626.1 JXUM01091236 JXUM01091237 KQ563898 KXJ73096.1 GFDL01008676 JAV26369.1 APGK01023663 KB740510 ENN80520.1 APCN01003750 AAAB01008984 EAA15110.3 AJWK01001704 AJWK01001705 AJWK01001706 AJWK01001707 GBYB01015092 JAG84859.1 JH432116 GEFH01003100 JAP65481.1 GEDC01029686 GEDC01022852 JAS07612.1 JAS14446.1 GFDF01001046 JAV13038.1 PCG80235.1 CVRI01000054 CRL00589.1 PZC77220.1 AJVK01002280 GANO01004231 JAB55640.1 ODYU01002281 SOQ39553.1 JO842597 AEO34214.1 GBBM01006655 JAC28763.1 LBMM01010836 KMQ87198.1 GEBQ01030844 GEBQ01003427 JAT09133.1 JAT36550.1 KQ978076 KYM97209.1 GFAC01007137 JAT92051.1 KQ971343 EFA04436.1 GANP01006687 JAB77781.1 NEVH01016296 PNF26063.1 KK119486 KFM75812.1 GBHO01041752 GBRD01009238 GDHC01021230 JAG01852.1 JAG56583.1 JAP97398.1 GECU01016971 JAS90735.1 KPJ13298.1 NNAY01000851 OXU26205.1 AK402950 BAM19475.1 GACK01006080 JAA58954.1 LRGB01000248 KZS20075.1 CAEY01000548 GFPF01006729 MAA17875.1 GEDV01006632 JAP81925.1 GEDV01010831 JAP77726.1 GDIQ01028464 JAN66273.1 ODYU01006327 SOQ48073.1 GECZ01027065 GECZ01021950 GECZ01016993 GECZ01014935 GECZ01002849 JAS42704.1 JAS47819.1 JAS52776.1 JAS54834.1 JAS66920.1 GALA01000709 JAA94143.1 GGLE01001088 MBY05214.1 DS231822 EDS25775.1 GBBI01003447 JAC15265.1
SOQ54698.1 NWSH01000047 PCG80234.1 AGBW02009824 OWR49838.1 KQ459584 KPI98453.1 JTDY01008744 KOB64425.1 KQ460644 KPJ13300.1 OWR49837.1 KPI98455.1 PZC77219.1 KQ976396 KYM92900.1 ADTU01011823 KQ982934 KYQ49150.1 PCG80236.1 GL888417 EGI61226.1 KQ978619 KYN29786.1 KQ981727 KYN37056.1 KPJ13299.1 GDAI01000074 JAI17529.1 KPI98454.1 GDQN01006658 JAT84396.1 GDQN01010932 GDQN01010586 GDQN01009981 GDQN01006844 GDQN01006638 GDQN01004581 GDQN01002001 JAT80122.1 JAT80468.1 JAT81073.1 JAT84210.1 JAT84416.1 JAT86473.1 JAT89053.1 GL447175 EFN86797.1 KZ288348 PBC27476.1 JR038487 AEY58275.1 AXCM01006387 GL443983 EFN61419.1 KQ435007 KZC13591.1 ADMH02000797 ETN65036.1 GGFM01007584 MBW28335.1 GGFK01011318 MBW44639.1 GALA01000040 JAA94812.1 KK107648 QOIP01000008 EZA48307.1 RLU19681.1 LJIG01022548 KRT79991.1 UFQT01000438 SSX24310.1 ATLV01022519 KE525333 KFB47728.1 KQ414598 KOC69815.1 CH477514 EAT39718.1 GAPW01004972 JAC08626.1 JXUM01091236 JXUM01091237 KQ563898 KXJ73096.1 GFDL01008676 JAV26369.1 APGK01023663 KB740510 ENN80520.1 APCN01003750 AAAB01008984 EAA15110.3 AJWK01001704 AJWK01001705 AJWK01001706 AJWK01001707 GBYB01015092 JAG84859.1 JH432116 GEFH01003100 JAP65481.1 GEDC01029686 GEDC01022852 JAS07612.1 JAS14446.1 GFDF01001046 JAV13038.1 PCG80235.1 CVRI01000054 CRL00589.1 PZC77220.1 AJVK01002280 GANO01004231 JAB55640.1 ODYU01002281 SOQ39553.1 JO842597 AEO34214.1 GBBM01006655 JAC28763.1 LBMM01010836 KMQ87198.1 GEBQ01030844 GEBQ01003427 JAT09133.1 JAT36550.1 KQ978076 KYM97209.1 GFAC01007137 JAT92051.1 KQ971343 EFA04436.1 GANP01006687 JAB77781.1 NEVH01016296 PNF26063.1 KK119486 KFM75812.1 GBHO01041752 GBRD01009238 GDHC01021230 JAG01852.1 JAG56583.1 JAP97398.1 GECU01016971 JAS90735.1 KPJ13298.1 NNAY01000851 OXU26205.1 AK402950 BAM19475.1 GACK01006080 JAA58954.1 LRGB01000248 KZS20075.1 CAEY01000548 GFPF01006729 MAA17875.1 GEDV01006632 JAP81925.1 GEDV01010831 JAP77726.1 GDIQ01028464 JAN66273.1 ODYU01006327 SOQ48073.1 GECZ01027065 GECZ01021950 GECZ01016993 GECZ01014935 GECZ01002849 JAS42704.1 JAS47819.1 JAS52776.1 JAS54834.1 JAS66920.1 GALA01000709 JAA94143.1 GGLE01001088 MBY05214.1 DS231822 EDS25775.1 GBBI01003447 JAC15265.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000053240
+ More
UP000192223 UP000078540 UP000005205 UP000075809 UP000007755 UP000078492 UP000078541 UP000069272 UP000008237 UP000242457 UP000075883 UP000000311 UP000076502 UP000000673 UP000075880 UP000053097 UP000279307 UP000030765 UP000005203 UP000075901 UP000053825 UP000075900 UP000008820 UP000069940 UP000249989 UP000076408 UP000075885 UP000019118 UP000075840 UP000075903 UP000076407 UP000007062 UP000092461 UP000075902 UP000075920 UP000183832 UP000092462 UP000036403 UP000078542 UP000007266 UP000235965 UP000054359 UP000215335 UP000076858 UP000015104 UP000002358 UP000002320
UP000192223 UP000078540 UP000005205 UP000075809 UP000007755 UP000078492 UP000078541 UP000069272 UP000008237 UP000242457 UP000075883 UP000000311 UP000076502 UP000000673 UP000075880 UP000053097 UP000279307 UP000030765 UP000005203 UP000075901 UP000053825 UP000075900 UP000008820 UP000069940 UP000249989 UP000076408 UP000075885 UP000019118 UP000075840 UP000075903 UP000076407 UP000007062 UP000092461 UP000075902 UP000075920 UP000183832 UP000092462 UP000036403 UP000078542 UP000007266 UP000235965 UP000054359 UP000215335 UP000076858 UP000015104 UP000002358 UP000002320
PRIDE
ProteinModelPortal
H9J999
A0A1E1W4N4
A0A2W1BSB3
A0A2H1WNT9
A0A2A4K8K1
A0A212F7Y8
+ More
A0A194PZI8 A0A0L7KMT7 A0A194R7T3 A0A212F7X2 A0A1W4W5Z0 A0A194Q4W7 A0A2W1BQ69 A0A195BX80 A0A158NCE3 A0A151WMV4 A0A2A4K8A6 F4WWY3 A0A151JQZ3 A0A195F9K1 A0A194R702 A0A182F5D6 A0A0K8TT38 A0A194Q0D3 A0A1E1WBQ1 A0A1E1W281 E2BBX3 A0A2A3E8E4 V9IB08 A0A182M1Q6 E2AYQ6 A0A154PNY3 W5JLN5 A0A2M3ZIN6 A0A2M4AV21 A0A182JF51 T1E3V1 A0A026VX22 A0A0T6AZD7 A0A336M2L0 A0A084WBY4 A0A087ZV46 A0A182T2H6 A0A0L7RG42 A0A182R9L1 Q16YL7 A0A023EI88 A0A182GVL0 A0A182YFB6 A0A182NZW2 A0A1Q3FFQ2 N6UFL6 A0A182I524 A0A182V9P4 A0A182WYP8 Q7PW02 A0A1B0C907 A0A0C9RNK5 T1JEB9 A0A182UCD8 A0A131XIV8 A0A1B6CLX8 A0A1L8E2T8 A0A182WH26 A0A2A4K7I5 A0A1J1IK14 A0A2W1BWN8 A0A1B0D093 U5EQI3 A0A2H1VFC6 G3ML46 A0A023G7D4 A0A0J7K9U5 A0A1B6KCF7 A0A151IBZ1 A0A1E1WZ53 D6WJ80 V5IGH3 A0A2J7QBU4 A0A087UEM3 A0A0A9W091 A0A1B6IUY1 A0A194R652 A0A232F6D9 I4DNI5 L7M764 A0A162QYT6 T1KTW6 A0A224YLQ2 A0A131YTR3 A0A131YI05 K7IZL1 A0A0P6IQR9 A0A2H1W4X8 A0A1B6EXG7 T1E2H4 A0A2R5L6S4 B0W1H7 A0A023F1B3
A0A194PZI8 A0A0L7KMT7 A0A194R7T3 A0A212F7X2 A0A1W4W5Z0 A0A194Q4W7 A0A2W1BQ69 A0A195BX80 A0A158NCE3 A0A151WMV4 A0A2A4K8A6 F4WWY3 A0A151JQZ3 A0A195F9K1 A0A194R702 A0A182F5D6 A0A0K8TT38 A0A194Q0D3 A0A1E1WBQ1 A0A1E1W281 E2BBX3 A0A2A3E8E4 V9IB08 A0A182M1Q6 E2AYQ6 A0A154PNY3 W5JLN5 A0A2M3ZIN6 A0A2M4AV21 A0A182JF51 T1E3V1 A0A026VX22 A0A0T6AZD7 A0A336M2L0 A0A084WBY4 A0A087ZV46 A0A182T2H6 A0A0L7RG42 A0A182R9L1 Q16YL7 A0A023EI88 A0A182GVL0 A0A182YFB6 A0A182NZW2 A0A1Q3FFQ2 N6UFL6 A0A182I524 A0A182V9P4 A0A182WYP8 Q7PW02 A0A1B0C907 A0A0C9RNK5 T1JEB9 A0A182UCD8 A0A131XIV8 A0A1B6CLX8 A0A1L8E2T8 A0A182WH26 A0A2A4K7I5 A0A1J1IK14 A0A2W1BWN8 A0A1B0D093 U5EQI3 A0A2H1VFC6 G3ML46 A0A023G7D4 A0A0J7K9U5 A0A1B6KCF7 A0A151IBZ1 A0A1E1WZ53 D6WJ80 V5IGH3 A0A2J7QBU4 A0A087UEM3 A0A0A9W091 A0A1B6IUY1 A0A194R652 A0A232F6D9 I4DNI5 L7M764 A0A162QYT6 T1KTW6 A0A224YLQ2 A0A131YTR3 A0A131YI05 K7IZL1 A0A0P6IQR9 A0A2H1W4X8 A0A1B6EXG7 T1E2H4 A0A2R5L6S4 B0W1H7 A0A023F1B3
Ontologies
GO
PANTHER
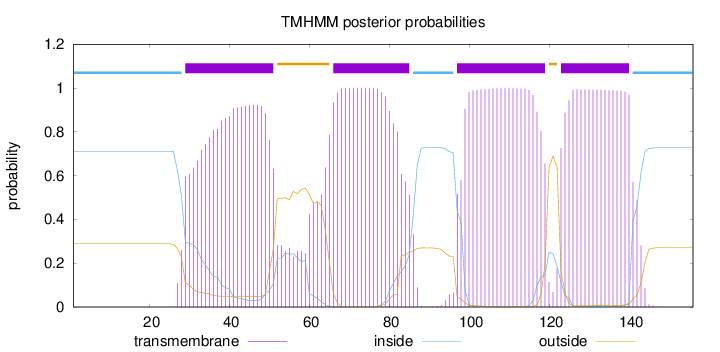
Topology
Length:
156
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
83.42897
Exp number, first 60 AAs:
21.43607
Total prob of N-in:
0.71148
POSSIBLE N-term signal
sequence
inside
1 - 28
TMhelix
29 - 51
outside
52 - 65
TMhelix
66 - 85
inside
86 - 96
TMhelix
97 - 119
outside
120 - 122
TMhelix
123 - 140
inside
141 - 156
Population Genetic Test Statistics
Pi
122.251302
Theta
115.225856
Tajima's D
-0.672882
CLR
1.837578
CSRT
0.201989900504975
Interpretation
Uncertain
