Gene
KWMTBOMO01613
Pre Gene Modal
BGIBMGA006090
Annotation
PREDICTED:_uncharacterized_protein_LOC101743230_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.195
Sequence
CDS
ATGGCTTCTAGTTATTTCACTGAAAGCGAGGCTCCGAGAGATTTTAGAGTTTCTTCAACAGAGAGTGACGGGCAATACACAAAATGCCACCTGTGCTATACAGTAAAGAGGAACTTTTACTGCACGGACTGTATAAAAGAAGGAAATTTTGTACATAGTTCAATGCCTTATTCGGACAGATATTCAGAAAAACTCTCAAAACTACTGAGATTAAAAATGAATCGCAAACACATACTAGATCGCTGTGAGAGGCTCCTAGCCCCGAAGCTAAAAAAGGACTCCCTCCTGACAGAAGCAAAACAATGTAGAGACAAAATTGACCTACTGAAATTAGCAATAAATCAAAGACGCAGCAATGTAGATGAAAAGAAAAAAGAGTTAAGTGAGCTAAAAACTTACAATACAGAATTGCGTTTACGGCTTCCAAGATATCAGAAAAGGGTAGCAAGTCTGGGTACCCATTCAGAGAAACAGAAATTGGAATTGCAGCATAAAATTAATCTGTACAATGAACAGGCTGACTCGTTGGCTGCACTACGCAGGTCTTGGATACGCCAACTGACCACTTATATATTTCCCGTGTATATGAGTTATGATACAAGTGACAGCATAGAAGACATGGAGTTTATAGGAGAAGACCTCCAAGAACCAGTGGGAAGAGCTCAACTACACATTGTTGCTCCAAGTATCAGCACTGATGGAGATTACTCGGATATAACCTTAGGGCCATTAAACAAGGAGGGCTCTACAACAGGAGTACCGGACGCGGCGGACAGCACCGCGGCGGCGCTGGGGCTGGCAGCACAGTTACTGTGCATGTTGGCCTGGATGTTACACATTCGCTTACCACATTCTTTATCATTGTCGGAGTACATGACGTCACGCGTGGAGTCGGCGGAGGCGTGCGTGAGCGCGCGGCGCGTGTGTGCGTGCGCGGGGGCGGTGTGTGCGGGAGCGGGGGTGCGCGCGGGGGCGGGGCAGGGGGGCGCGCTGCACTCGCTGCACGCTCTGCACGCACTGGCGCTCGCCGCCGCCGTGGACGACCCGCTGCTGGCCAGGTAA
Protein
MASSYFTESEAPRDFRVSSTESDGQYTKCHLCYTVKRNFYCTDCIKEGNFVHSSMPYSDRYSEKLSKLLRLKMNRKHILDRCERLLAPKLKKDSLLTEAKQCRDKIDLLKLAINQRRSNVDEKKKELSELKTYNTELRLRLPRYQKRVASLGTHSEKQKLELQHKINLYNEQADSLAALRRSWIRQLTTYIFPVYMSYDTSDSIEDMEFIGEDLQEPVGRAQLHIVAPSISTDGDYSDITLGPLNKEGSTTGVPDAADSTAAALGLAAQLLCMLAWMLHIRLPHSLSLSEYMTSRVESAEACVSARRVCACAGAVCAGAGVRAGAGQGGALHSLHALHALALAAAVDDPLLAR
Summary
Uniprot
A0A1E1W3G5
A0A2H1W0I9
A0A212F7X6
A0A194Q4W2
A0A194R658
A0A0L7K4W0
+ More
A0A0A9Z2G3 A0A146MBZ5 A0A154PH51 A0A0T6BDK8 A0A158NC82 A0A087ZQ93 A0A0N0BH92 A0A151HZS6 A0A067RAQ4 A0A0K8SSI9 E1ZVH6 A0A3L8D412 A0A026X1S0 E9ILY0 A0A232ETE6 A0A195EAC2 A0A195CGF5 A0A195FAS9 A0A2J7RRQ8 A0A0L7R4F8 A0A2A3EJR3 A0A023EZ64 A0A310S9T9 A0A0V0GD05 A0A0C9R654 A0A182G658 A0A1S4EXW8 Q17M47 A0A336KK69 A0A336MIG2 A0A1Y1NGI8 A0A1Y1NFW9 A0A1Q3FAY4 A0A1Q3FAQ7 A0A1Q3FA33 B0WEJ7 A0A0C9R0M9 A0A182FUF3 A0A2R7W1Z4 A0A2M4AJS3 A0A2M4AJS7 A0A084WB44 A0A2M4AK29 W5J3N4 T1PIH9 A0A1S3K0L6 B4NBF8 A0A1S3HR01 A0A182NPG2 A0A1S3HPK1 A0A182JWG4 A0A1I8P185 A0A182Q0G2 A0A0M4ELZ1 A0A182PBM9
A0A0A9Z2G3 A0A146MBZ5 A0A154PH51 A0A0T6BDK8 A0A158NC82 A0A087ZQ93 A0A0N0BH92 A0A151HZS6 A0A067RAQ4 A0A0K8SSI9 E1ZVH6 A0A3L8D412 A0A026X1S0 E9ILY0 A0A232ETE6 A0A195EAC2 A0A195CGF5 A0A195FAS9 A0A2J7RRQ8 A0A0L7R4F8 A0A2A3EJR3 A0A023EZ64 A0A310S9T9 A0A0V0GD05 A0A0C9R654 A0A182G658 A0A1S4EXW8 Q17M47 A0A336KK69 A0A336MIG2 A0A1Y1NGI8 A0A1Y1NFW9 A0A1Q3FAY4 A0A1Q3FAQ7 A0A1Q3FA33 B0WEJ7 A0A0C9R0M9 A0A182FUF3 A0A2R7W1Z4 A0A2M4AJS3 A0A2M4AJS7 A0A084WB44 A0A2M4AK29 W5J3N4 T1PIH9 A0A1S3K0L6 B4NBF8 A0A1S3HR01 A0A182NPG2 A0A1S3HPK1 A0A182JWG4 A0A1I8P185 A0A182Q0G2 A0A0M4ELZ1 A0A182PBM9
Pubmed
EMBL
GDQN01009527
JAT81527.1
ODYU01005302
SOQ46024.1
AGBW02009824
OWR49842.1
+ More
KQ459584 KPI98450.1 KQ460644 KPJ13303.1 JTDY01010643 KOB58136.1 GBHO01006066 JAG37538.1 GDHC01002302 JAQ16327.1 KQ434904 KZC11185.1 LJIG01001520 KRT85420.1 ADTU01011737 KQ435756 KOX75892.1 KQ976642 KYM78692.1 KK852577 KDR20930.1 GBRD01009497 JAG56327.1 GL434534 EFN74813.1 QOIP01000014 RLU14743.1 KK107061 EZA61339.1 GL764129 EFZ18518.1 NNAY01002288 OXU21614.1 KQ979236 KYN22163.1 KQ977873 KYM99168.1 KQ981727 KYN37144.1 NEVH01000599 PNF43516.1 KQ414658 KOC65641.1 KZ288229 PBC31714.1 GBBI01004212 JAC14500.1 KQ767768 OAD53311.1 GECL01000871 JAP05253.1 GBYB01011764 JAG81531.1 JXUM01000630 JXUM01000631 JXUM01000632 KQ560109 KXJ84470.1 CH477208 EAT47795.1 UFQS01000555 UFQT01000555 SSX04889.1 SSX25252.1 UFQS01001272 UFQT01001272 SSX10073.1 SSX29795.1 GEZM01003635 JAV96668.1 GEZM01003634 JAV96669.1 GFDL01010314 JAV24731.1 GFDL01010389 JAV24656.1 GFDL01010609 JAV24436.1 DS231910 EDS45713.1 GBYB01009694 JAG79461.1 KK854240 PTY13599.1 GGFK01007700 MBW41021.1 GGFK01007718 MBW41039.1 ATLV01022312 KE525331 KFB47438.1 GGFK01007786 MBW41107.1 ADMH02002188 ETN57913.1 KA648566 AFP63195.1 CH964232 EDW81122.2 AXCN02001828 CP012526 ALC46402.1
KQ459584 KPI98450.1 KQ460644 KPJ13303.1 JTDY01010643 KOB58136.1 GBHO01006066 JAG37538.1 GDHC01002302 JAQ16327.1 KQ434904 KZC11185.1 LJIG01001520 KRT85420.1 ADTU01011737 KQ435756 KOX75892.1 KQ976642 KYM78692.1 KK852577 KDR20930.1 GBRD01009497 JAG56327.1 GL434534 EFN74813.1 QOIP01000014 RLU14743.1 KK107061 EZA61339.1 GL764129 EFZ18518.1 NNAY01002288 OXU21614.1 KQ979236 KYN22163.1 KQ977873 KYM99168.1 KQ981727 KYN37144.1 NEVH01000599 PNF43516.1 KQ414658 KOC65641.1 KZ288229 PBC31714.1 GBBI01004212 JAC14500.1 KQ767768 OAD53311.1 GECL01000871 JAP05253.1 GBYB01011764 JAG81531.1 JXUM01000630 JXUM01000631 JXUM01000632 KQ560109 KXJ84470.1 CH477208 EAT47795.1 UFQS01000555 UFQT01000555 SSX04889.1 SSX25252.1 UFQS01001272 UFQT01001272 SSX10073.1 SSX29795.1 GEZM01003635 JAV96668.1 GEZM01003634 JAV96669.1 GFDL01010314 JAV24731.1 GFDL01010389 JAV24656.1 GFDL01010609 JAV24436.1 DS231910 EDS45713.1 GBYB01009694 JAG79461.1 KK854240 PTY13599.1 GGFK01007700 MBW41021.1 GGFK01007718 MBW41039.1 ATLV01022312 KE525331 KFB47438.1 GGFK01007786 MBW41107.1 ADMH02002188 ETN57913.1 KA648566 AFP63195.1 CH964232 EDW81122.2 AXCN02001828 CP012526 ALC46402.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000037510
UP000076502
UP000005205
+ More
UP000005203 UP000053105 UP000078540 UP000027135 UP000000311 UP000279307 UP000053097 UP000215335 UP000078492 UP000078542 UP000078541 UP000235965 UP000053825 UP000242457 UP000069940 UP000249989 UP000008820 UP000002320 UP000069272 UP000030765 UP000000673 UP000095301 UP000085678 UP000007798 UP000075884 UP000075881 UP000095300 UP000075886 UP000092553 UP000075885
UP000005203 UP000053105 UP000078540 UP000027135 UP000000311 UP000279307 UP000053097 UP000215335 UP000078492 UP000078542 UP000078541 UP000235965 UP000053825 UP000242457 UP000069940 UP000249989 UP000008820 UP000002320 UP000069272 UP000030765 UP000000673 UP000095301 UP000085678 UP000007798 UP000075884 UP000075881 UP000095300 UP000075886 UP000092553 UP000075885
Pfam
PF10186 Atg14
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
A0A1E1W3G5
A0A2H1W0I9
A0A212F7X6
A0A194Q4W2
A0A194R658
A0A0L7K4W0
+ More
A0A0A9Z2G3 A0A146MBZ5 A0A154PH51 A0A0T6BDK8 A0A158NC82 A0A087ZQ93 A0A0N0BH92 A0A151HZS6 A0A067RAQ4 A0A0K8SSI9 E1ZVH6 A0A3L8D412 A0A026X1S0 E9ILY0 A0A232ETE6 A0A195EAC2 A0A195CGF5 A0A195FAS9 A0A2J7RRQ8 A0A0L7R4F8 A0A2A3EJR3 A0A023EZ64 A0A310S9T9 A0A0V0GD05 A0A0C9R654 A0A182G658 A0A1S4EXW8 Q17M47 A0A336KK69 A0A336MIG2 A0A1Y1NGI8 A0A1Y1NFW9 A0A1Q3FAY4 A0A1Q3FAQ7 A0A1Q3FA33 B0WEJ7 A0A0C9R0M9 A0A182FUF3 A0A2R7W1Z4 A0A2M4AJS3 A0A2M4AJS7 A0A084WB44 A0A2M4AK29 W5J3N4 T1PIH9 A0A1S3K0L6 B4NBF8 A0A1S3HR01 A0A182NPG2 A0A1S3HPK1 A0A182JWG4 A0A1I8P185 A0A182Q0G2 A0A0M4ELZ1 A0A182PBM9
A0A0A9Z2G3 A0A146MBZ5 A0A154PH51 A0A0T6BDK8 A0A158NC82 A0A087ZQ93 A0A0N0BH92 A0A151HZS6 A0A067RAQ4 A0A0K8SSI9 E1ZVH6 A0A3L8D412 A0A026X1S0 E9ILY0 A0A232ETE6 A0A195EAC2 A0A195CGF5 A0A195FAS9 A0A2J7RRQ8 A0A0L7R4F8 A0A2A3EJR3 A0A023EZ64 A0A310S9T9 A0A0V0GD05 A0A0C9R654 A0A182G658 A0A1S4EXW8 Q17M47 A0A336KK69 A0A336MIG2 A0A1Y1NGI8 A0A1Y1NFW9 A0A1Q3FAY4 A0A1Q3FAQ7 A0A1Q3FA33 B0WEJ7 A0A0C9R0M9 A0A182FUF3 A0A2R7W1Z4 A0A2M4AJS3 A0A2M4AJS7 A0A084WB44 A0A2M4AK29 W5J3N4 T1PIH9 A0A1S3K0L6 B4NBF8 A0A1S3HR01 A0A182NPG2 A0A1S3HPK1 A0A182JWG4 A0A1I8P185 A0A182Q0G2 A0A0M4ELZ1 A0A182PBM9
Ontologies
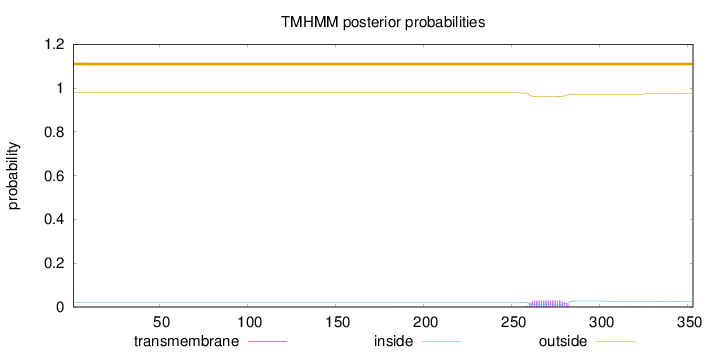
Topology
Length:
353
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.71512
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02129
outside
1 - 353
Population Genetic Test Statistics
Pi
210.333471
Theta
170.509173
Tajima's D
0.870068
CLR
0.284362
CSRT
0.623818809059547
Interpretation
Uncertain
