Gene
KWMTBOMO01610
Pre Gene Modal
BGIBMGA006086
Annotation
hypothetical_protein_KGM_11415_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 3.559
Sequence
CDS
ATGGATATTTCGTATCAGTATTCTAAGAAACGATGCCGTTTCGGTGGTCAGCCTCAGTTCTGCGAGCAGGGCCCGGAGATGTGCGACAGCATCGCCACGAATGTCGTCGAGCACAAGAACTTCATCCTGCGGAACCCCGTGCACCAGCCCACGCAGCACACTCCTTTCTATTCTGAAAACAACGCTAATACCATCAGAACGGACTACAGCTCATCAGGAGCGAACCACGCGGAAGGTGGGTGGCCTAAAGATGTCAGCGTCATCGATCCCGAAGCCACACAGCGGTACCGACGTAAAGTTGAAAAAGATGACAACTACATCAACTGCGTCATGTCCACTGCTCCGGAGATGGATCATTACGTGCTCCAAAACAACGCCATCGACATGTACCGAACGTACTATGCAGAAATGCCATCTATGCCAGAACTGGAACGCATCAGCTGTCGCACTGTGAACGTGTATCGCGACGGTGATGGAACGGAAGCGACGCGACCGGTCAGCTCCATATGCTGGCAGCCTGATGGCGGCCAACGCTTCGCAGTCACCTACATCGACGTGGACTTCAACCGCAACGCGCGAGCGCCCATTGTCTCTTACATTTGGAACTTGGAGAACGCGAATCAACCAGAAGTAACGTTCACACCACCATGCCCGCTCTTAGATTTGATATATAATCCTCGTGATCAAAACCTTATGGCTGGTGGTTTGGTTAATGGACAGGTCGGCGTGTGGGACGCACGAAAAGGCGGCGATCCAACTCTCACATGTCCACCGCATGTTGCCCATCGAGATCTTGTGCGAAACGTCATATTCATCAACTCTAAATCGGGACTAGAGTTCTTCTCTGGAGGATCAGACGGGGCCTGTAAGTGGTGGGACATCCGAAACATGAGTGAGCCTGTGGAAGAGATGATTTTAGATGCAGTGAAGTCTTCATTTGACGTTCAAACGATGGCTAAAGCTAACGGTGTGACAGCATTAGAATTCGAGCCCACTATGCCGACGCGCTTCATGGCTGGAACTGAAAACGGACTTATCATCGGAGGCAACCGTAAAGGAAAGACACCGTTAGAGAAAATACCTCTTAAGATGGAAGCGCACCTGGGTCCCATCTGGTCCCTGGAGCGTAATCCGGGATTTTTGAAAAATTTTCTAACCGTTGGAGATTGGACAGCACGAATTTGGAGCGAGGACTGTCGCGAATCTCCCGTACTATGGTCTCCTCTACATCGGCACAAGATCACTGCTGGTACTTGGAGTCCAACTAGGTCCTCATTGATGCTGCTGGCTCAGTGGAACGGGACGCTAGCGACGTGGGACCTGTTGCGGCGTCAGCATGAGCCCGTACTCACTATGCACGTCTGTGATGAACCGCTTTTGAGGCTCAAAACTCATGAAAGGGGTACGCTCGTGGCTTGCGGGAGTCAGAAGGGTAGCATTTTCATGGTGGAACTGTCGCAAACCTTGACACAGTCTGATAAAAACGATAAGACATTGCTGACAGCGGTATTGGAACGGGAGAGTAAAAGGGAGCGTATTCTGGAGGCTCGCATGCGTGAGATTAGATTGAAGATGCGTCAGGCCGAGGAGGGCTCGCCGCGCGCCTCGCTCACGGAGCCCGAGCTACCCTTTGACGTCAACGCTGTCACGGCCGAGTACTTTGCAATCGTGAAGAAAGAACTAGCCGCCCTGTGA
Protein
MDISYQYSKKRCRFGGQPQFCEQGPEMCDSIATNVVEHKNFILRNPVHQPTQHTPFYSENNANTIRTDYSSSGANHAEGGWPKDVSVIDPEATQRYRRKVEKDDNYINCVMSTAPEMDHYVLQNNAIDMYRTYYAEMPSMPELERISCRTVNVYRDGDGTEATRPVSSICWQPDGGQRFAVTYIDVDFNRNARAPIVSYIWNLENANQPEVTFTPPCPLLDLIYNPRDQNLMAGGLVNGQVGVWDARKGGDPTLTCPPHVAHRDLVRNVIFINSKSGLEFFSGGSDGACKWWDIRNMSEPVEEMILDAVKSSFDVQTMAKANGVTALEFEPTMPTRFMAGTENGLIIGGNRKGKTPLEKIPLKMEAHLGPIWSLERNPGFLKNFLTVGDWTARIWSEDCRESPVLWSPLHRHKITAGTWSPTRSSLMLLAQWNGTLATWDLLRRQHEPVLTMHVCDEPLLRLKTHERGTLVACGSQKGSIFMVELSQTLTQSDKNDKTLLTAVLERESKRERILEARMREIRLKMRQAEEGSPRASLTEPELPFDVNAVTAEYFAIVKKELAAL
Summary
Uniprot
H9J994
H9J995
A0A212F7F5
A0A2W1BCK5
A0A2H1V4H2
A0A0L7KHE8
+ More
A0A194QXZ9 A0A0L7KUC8 A0A2A4JFG7 A0A2W1BBN8 A0A194PF73 A0A2H1V2V5 A0A194PLU7 A0A2H1W9I7 A0A194QZY6 A0A2A4JH19 A0A212FGV3 H9JQE6 A0A2A4K9W5 A0A1B6H019 E0VIU6 A0A067QXI2 D6WDC8 B4KUS4 A0A0Q9XLF0 A0A1I8NBK1 B4J2I6 A0A0Q9WXR2 B4LCQ9 A0A0R3P4Z7 B4HAH2 B3M4B0 Q2LZ77 A0A0L0BL07 F4W6B8 A0A1W4VTJ2 A0A1B0B9U4 A0A1A9VG63 E2BG01 A0A3B0JVL1 A0A158P2L6 A0A0M4EBX6 A0A0Q9WSC5 A0A1A9WH63 Q4V560 B4N4L9 M9PC80 Q9VTM3 B4QQ99 B4HEX4 B4PFM9 A0A195FJF9 B3NGZ3 A0A1Y1KE26 A0A1Y1K6N5 A0A151XCS7 A0A2J7QWB5 N6T1P0 B0WLC7 A0A3L8DB42 Q7QGH2 A0A182ITQ6 A0A1Q3FTJ5 A0A182XC21 A0A182V9I7 A0A182LP72 A0A182HNC0 A0A182MUH2 A0A182WEV4 A0A195DMF1 A0A0J9UJB0 A0A0J9RUU5 A0A336LZL3 A0A336MZX5 E2B285 A0A182MY67 A0A182KAP0 A0A0L7QZ97 A0A182Y5Y0 Q16TL6 A0A182R0Y4 A0A1J1HMN4 A0A1B0C9D7 A0A026VZA0 A0A1B0G3X0 A0A182RD31 A0A182FC30 W5JAU1 A0A088A7L4 A0A154PC68 K7IM39 A0A210PH44 A0A1B0D579 A0A3P8YL09 A0A182TS26 A0A0N0BC27 A0A3B4DKT8 V4B996 A0A2D0S6C3 F6XSE5
A0A194QXZ9 A0A0L7KUC8 A0A2A4JFG7 A0A2W1BBN8 A0A194PF73 A0A2H1V2V5 A0A194PLU7 A0A2H1W9I7 A0A194QZY6 A0A2A4JH19 A0A212FGV3 H9JQE6 A0A2A4K9W5 A0A1B6H019 E0VIU6 A0A067QXI2 D6WDC8 B4KUS4 A0A0Q9XLF0 A0A1I8NBK1 B4J2I6 A0A0Q9WXR2 B4LCQ9 A0A0R3P4Z7 B4HAH2 B3M4B0 Q2LZ77 A0A0L0BL07 F4W6B8 A0A1W4VTJ2 A0A1B0B9U4 A0A1A9VG63 E2BG01 A0A3B0JVL1 A0A158P2L6 A0A0M4EBX6 A0A0Q9WSC5 A0A1A9WH63 Q4V560 B4N4L9 M9PC80 Q9VTM3 B4QQ99 B4HEX4 B4PFM9 A0A195FJF9 B3NGZ3 A0A1Y1KE26 A0A1Y1K6N5 A0A151XCS7 A0A2J7QWB5 N6T1P0 B0WLC7 A0A3L8DB42 Q7QGH2 A0A182ITQ6 A0A1Q3FTJ5 A0A182XC21 A0A182V9I7 A0A182LP72 A0A182HNC0 A0A182MUH2 A0A182WEV4 A0A195DMF1 A0A0J9UJB0 A0A0J9RUU5 A0A336LZL3 A0A336MZX5 E2B285 A0A182MY67 A0A182KAP0 A0A0L7QZ97 A0A182Y5Y0 Q16TL6 A0A182R0Y4 A0A1J1HMN4 A0A1B0C9D7 A0A026VZA0 A0A1B0G3X0 A0A182RD31 A0A182FC30 W5JAU1 A0A088A7L4 A0A154PC68 K7IM39 A0A210PH44 A0A1B0D579 A0A3P8YL09 A0A182TS26 A0A0N0BC27 A0A3B4DKT8 V4B996 A0A2D0S6C3 F6XSE5
Pubmed
19121390
22118469
28756777
26227816
26354079
20566863
+ More
24845553 18362917 19820115 17994087 25315136 15632085 26108605 21719571 20798317 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 28004739 23537049 30249741 12364791 20966253 22936249 25244985 17510324 24508170 20920257 23761445 20075255 28812685 25069045 23254933 20431018
24845553 18362917 19820115 17994087 25315136 15632085 26108605 21719571 20798317 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 28004739 23537049 30249741 12364791 20966253 22936249 25244985 17510324 24508170 20920257 23761445 20075255 28812685 25069045 23254933 20431018
EMBL
BABH01004332
AGBW02009873
OWR49674.1
KZ150474
PZC70746.1
ODYU01000427
+ More
SOQ35174.1 JTDY01009838 KOB62521.1 KQ461154 KPJ08451.1 JTDY01005576 KOB66868.1 NWSH01001606 PCG70721.1 PZC70747.1 KQ459606 KPI91673.1 SOQ35175.1 KQ459599 KPI94411.1 ODYU01007193 SOQ49759.1 KQ460883 KPJ11108.1 PCG70720.1 AGBW02008593 OWR52965.1 BABH01011924 BABH01011925 BABH01011926 NWSH01000021 PCG80688.1 GECZ01001756 JAS68013.1 DS235206 EEB13302.1 KK852855 KDR14924.1 KQ971321 EEZ99548.1 CH933809 EDW19330.1 KRG06602.1 CH916366 EDV97071.1 CH940647 KRF85205.1 EDW70951.1 CH379069 KRT08217.1 CH479240 EDW37570.1 CH902618 EDV39380.2 EAL29631.2 JRES01001711 KNC20623.1 GL887707 EGI70253.1 JXJN01010587 GL448096 EFN85403.1 OUUW01000002 SPP76771.1 ADTU01006976 CP012525 ALC45054.1 CH964095 KRF99008.1 BT022764 BT022796 BT022804 AAY55180.1 AAY55212.1 AAY55220.1 EDW79093.1 AE014296 AGB94417.1 AAF50024.2 CM000363 EDX10117.1 CH480815 EDW41143.1 CM000159 EDW94178.2 KQ981523 KYN40397.1 CH954178 EDV51450.2 GEZM01091218 JAV57017.1 GEZM01091208 JAV57034.1 KQ982297 KYQ58174.1 NEVH01009765 PNF32875.1 APGK01055684 APGK01055685 KB741269 KB632330 ENN71438.1 ERL92589.1 DS231984 EDS30420.1 QOIP01000010 RLU17333.1 AAAB01008834 EAA05710.4 GFDL01004302 JAV30743.1 APCN01001903 AXCM01003863 KQ980724 KYN14063.1 CM002912 KMY99050.1 KMY99049.1 UFQT01000346 SSX23434.1 UFQT01002710 SSX33977.1 GL445067 EFN60211.1 KQ414681 KOC63945.1 CH477645 EAT37861.1 AXCN02002036 CVRI01000011 CRK89309.1 AJWK01002233 AJWK01002234 AJWK01002235 AJWK01002236 KK107670 EZA48184.1 CCAG010011538 ADMH02001775 ETN61121.1 KQ434870 KZC09485.1 NEDP02076708 OWF35787.1 AJVK01003394 AJVK01003395 KQ435955 KOX68072.1 KB199905 ESP03921.1 AAMC01045484
SOQ35174.1 JTDY01009838 KOB62521.1 KQ461154 KPJ08451.1 JTDY01005576 KOB66868.1 NWSH01001606 PCG70721.1 PZC70747.1 KQ459606 KPI91673.1 SOQ35175.1 KQ459599 KPI94411.1 ODYU01007193 SOQ49759.1 KQ460883 KPJ11108.1 PCG70720.1 AGBW02008593 OWR52965.1 BABH01011924 BABH01011925 BABH01011926 NWSH01000021 PCG80688.1 GECZ01001756 JAS68013.1 DS235206 EEB13302.1 KK852855 KDR14924.1 KQ971321 EEZ99548.1 CH933809 EDW19330.1 KRG06602.1 CH916366 EDV97071.1 CH940647 KRF85205.1 EDW70951.1 CH379069 KRT08217.1 CH479240 EDW37570.1 CH902618 EDV39380.2 EAL29631.2 JRES01001711 KNC20623.1 GL887707 EGI70253.1 JXJN01010587 GL448096 EFN85403.1 OUUW01000002 SPP76771.1 ADTU01006976 CP012525 ALC45054.1 CH964095 KRF99008.1 BT022764 BT022796 BT022804 AAY55180.1 AAY55212.1 AAY55220.1 EDW79093.1 AE014296 AGB94417.1 AAF50024.2 CM000363 EDX10117.1 CH480815 EDW41143.1 CM000159 EDW94178.2 KQ981523 KYN40397.1 CH954178 EDV51450.2 GEZM01091218 JAV57017.1 GEZM01091208 JAV57034.1 KQ982297 KYQ58174.1 NEVH01009765 PNF32875.1 APGK01055684 APGK01055685 KB741269 KB632330 ENN71438.1 ERL92589.1 DS231984 EDS30420.1 QOIP01000010 RLU17333.1 AAAB01008834 EAA05710.4 GFDL01004302 JAV30743.1 APCN01001903 AXCM01003863 KQ980724 KYN14063.1 CM002912 KMY99050.1 KMY99049.1 UFQT01000346 SSX23434.1 UFQT01002710 SSX33977.1 GL445067 EFN60211.1 KQ414681 KOC63945.1 CH477645 EAT37861.1 AXCN02002036 CVRI01000011 CRK89309.1 AJWK01002233 AJWK01002234 AJWK01002235 AJWK01002236 KK107670 EZA48184.1 CCAG010011538 ADMH02001775 ETN61121.1 KQ434870 KZC09485.1 NEDP02076708 OWF35787.1 AJVK01003394 AJVK01003395 KQ435955 KOX68072.1 KB199905 ESP03921.1 AAMC01045484
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000218220
UP000053268
+ More
UP000009046 UP000027135 UP000007266 UP000009192 UP000095301 UP000001070 UP000008792 UP000001819 UP000008744 UP000007801 UP000037069 UP000007755 UP000192221 UP000092460 UP000078200 UP000008237 UP000268350 UP000005205 UP000092553 UP000007798 UP000091820 UP000000803 UP000000304 UP000001292 UP000002282 UP000078541 UP000008711 UP000075809 UP000235965 UP000019118 UP000030742 UP000002320 UP000279307 UP000007062 UP000075880 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075920 UP000078492 UP000000311 UP000075884 UP000075881 UP000053825 UP000076408 UP000008820 UP000075886 UP000183832 UP000092461 UP000053097 UP000092444 UP000075900 UP000069272 UP000000673 UP000005203 UP000076502 UP000002358 UP000242188 UP000092462 UP000265140 UP000075902 UP000053105 UP000261440 UP000030746 UP000221080 UP000008143
UP000009046 UP000027135 UP000007266 UP000009192 UP000095301 UP000001070 UP000008792 UP000001819 UP000008744 UP000007801 UP000037069 UP000007755 UP000192221 UP000092460 UP000078200 UP000008237 UP000268350 UP000005205 UP000092553 UP000007798 UP000091820 UP000000803 UP000000304 UP000001292 UP000002282 UP000078541 UP000008711 UP000075809 UP000235965 UP000019118 UP000030742 UP000002320 UP000279307 UP000007062 UP000075880 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075920 UP000078492 UP000000311 UP000075884 UP000075881 UP000053825 UP000076408 UP000008820 UP000075886 UP000183832 UP000092461 UP000053097 UP000092444 UP000075900 UP000069272 UP000000673 UP000005203 UP000076502 UP000002358 UP000242188 UP000092462 UP000265140 UP000075902 UP000053105 UP000261440 UP000030746 UP000221080 UP000008143
Interpro
Gene 3D
ProteinModelPortal
H9J994
H9J995
A0A212F7F5
A0A2W1BCK5
A0A2H1V4H2
A0A0L7KHE8
+ More
A0A194QXZ9 A0A0L7KUC8 A0A2A4JFG7 A0A2W1BBN8 A0A194PF73 A0A2H1V2V5 A0A194PLU7 A0A2H1W9I7 A0A194QZY6 A0A2A4JH19 A0A212FGV3 H9JQE6 A0A2A4K9W5 A0A1B6H019 E0VIU6 A0A067QXI2 D6WDC8 B4KUS4 A0A0Q9XLF0 A0A1I8NBK1 B4J2I6 A0A0Q9WXR2 B4LCQ9 A0A0R3P4Z7 B4HAH2 B3M4B0 Q2LZ77 A0A0L0BL07 F4W6B8 A0A1W4VTJ2 A0A1B0B9U4 A0A1A9VG63 E2BG01 A0A3B0JVL1 A0A158P2L6 A0A0M4EBX6 A0A0Q9WSC5 A0A1A9WH63 Q4V560 B4N4L9 M9PC80 Q9VTM3 B4QQ99 B4HEX4 B4PFM9 A0A195FJF9 B3NGZ3 A0A1Y1KE26 A0A1Y1K6N5 A0A151XCS7 A0A2J7QWB5 N6T1P0 B0WLC7 A0A3L8DB42 Q7QGH2 A0A182ITQ6 A0A1Q3FTJ5 A0A182XC21 A0A182V9I7 A0A182LP72 A0A182HNC0 A0A182MUH2 A0A182WEV4 A0A195DMF1 A0A0J9UJB0 A0A0J9RUU5 A0A336LZL3 A0A336MZX5 E2B285 A0A182MY67 A0A182KAP0 A0A0L7QZ97 A0A182Y5Y0 Q16TL6 A0A182R0Y4 A0A1J1HMN4 A0A1B0C9D7 A0A026VZA0 A0A1B0G3X0 A0A182RD31 A0A182FC30 W5JAU1 A0A088A7L4 A0A154PC68 K7IM39 A0A210PH44 A0A1B0D579 A0A3P8YL09 A0A182TS26 A0A0N0BC27 A0A3B4DKT8 V4B996 A0A2D0S6C3 F6XSE5
A0A194QXZ9 A0A0L7KUC8 A0A2A4JFG7 A0A2W1BBN8 A0A194PF73 A0A2H1V2V5 A0A194PLU7 A0A2H1W9I7 A0A194QZY6 A0A2A4JH19 A0A212FGV3 H9JQE6 A0A2A4K9W5 A0A1B6H019 E0VIU6 A0A067QXI2 D6WDC8 B4KUS4 A0A0Q9XLF0 A0A1I8NBK1 B4J2I6 A0A0Q9WXR2 B4LCQ9 A0A0R3P4Z7 B4HAH2 B3M4B0 Q2LZ77 A0A0L0BL07 F4W6B8 A0A1W4VTJ2 A0A1B0B9U4 A0A1A9VG63 E2BG01 A0A3B0JVL1 A0A158P2L6 A0A0M4EBX6 A0A0Q9WSC5 A0A1A9WH63 Q4V560 B4N4L9 M9PC80 Q9VTM3 B4QQ99 B4HEX4 B4PFM9 A0A195FJF9 B3NGZ3 A0A1Y1KE26 A0A1Y1K6N5 A0A151XCS7 A0A2J7QWB5 N6T1P0 B0WLC7 A0A3L8DB42 Q7QGH2 A0A182ITQ6 A0A1Q3FTJ5 A0A182XC21 A0A182V9I7 A0A182LP72 A0A182HNC0 A0A182MUH2 A0A182WEV4 A0A195DMF1 A0A0J9UJB0 A0A0J9RUU5 A0A336LZL3 A0A336MZX5 E2B285 A0A182MY67 A0A182KAP0 A0A0L7QZ97 A0A182Y5Y0 Q16TL6 A0A182R0Y4 A0A1J1HMN4 A0A1B0C9D7 A0A026VZA0 A0A1B0G3X0 A0A182RD31 A0A182FC30 W5JAU1 A0A088A7L4 A0A154PC68 K7IM39 A0A210PH44 A0A1B0D579 A0A3P8YL09 A0A182TS26 A0A0N0BC27 A0A3B4DKT8 V4B996 A0A2D0S6C3 F6XSE5
PDB
6F3A
E-value=2.79282e-16,
Score=210
Ontologies
GO
Topology
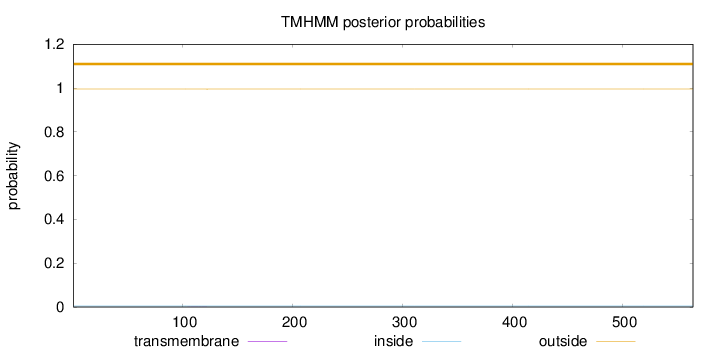
Length:
564
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00383
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00473
outside
1 - 564
Population Genetic Test Statistics
Pi
282.262654
Theta
204.624551
Tajima's D
1.050332
CLR
0.467135
CSRT
0.670716464176791
Interpretation
Uncertain
