Gene
KWMTBOMO01609
Pre Gene Modal
BGIBMGA006081
Annotation
antennal_esterase_precursor_[Bombyx_mori]
Full name
Carboxylic ester hydrolase
Location in the cell
Nuclear Reliability : 4.038
Sequence
CDS
ATGTGCGTGTCAATAAAGTGGGTCGTGCTGTGGAGTCTTTGGGCGGGAAGGCTGGTGCGGCAACCGACTCCGGCAGTGAGACTGAGTAACCATCGGCTCGTCAGCGGGCAGGTTGCCCCGGACGGGTCACATGTTCAATATTTTGGGATACCTTATGCTACTGTTCGACGTAGACACAGGTTCAAGGCAGCAGGCCTAGAGCCGGAATGGGATGGTGTGTTCAATGCTGTAAATGAACATGTGAGGTGTCCACAAAAATTTGCTGACAACTTTGTTTGGGGCCAAGAGGATTGCCTCATTATAAATATTTTCCATCCACTTTACTCAGAAGTCGATAAACTTTTACCGGTGAAAGTTTTCATCCATGGAGGTGGATTCCGTGAAGGATCATCGTCGAGACTTCTATTTGGCCCAGATTACCTGGTCCGTAAGAATATTGTCTTAGTGACCTTTAACTACCGAGTAGCCGTAACTGGGTTTTTGTGTCTCGGAATCAAAGATGCTCCAGGAAATGCAGGATTGAAAGATCAGGTTGCAGCACTGAAGTGGGTTCAACGAAATATCAAAGTTTTTGGCGGCGATCCTGACAATGTGACGATTTTCGGAGAAAGCGCCGGTGCTGCGTCTGTTTCTTTACATATCGTTTCACCAGCTTCTAAAGGTCTTTTCCATAAGGCTATCCTCCAAAGTGGGTCGTCGATATCACCATGGGCTGTCGAATTCGAACCAATTAAATATGCTAGCAGAGTAGCTGAAGTCTTAGGATATAAAACCATGGATCCGTATCAACTTTATGACATTCATATGAACCACACCATAGAAAGTATTGTGGGCGCGCCCGTCCCATTACTAGAAAGTCAAGTAGCAGCAGCTCAGCTAATTAATTCTCCCTGTATTGAAAAATCACACCCTGGTGTGGAGCCATTTCTTCTAGAATCGCCATTCGATATTTTTACCAGAGGTGAATACAACAAAGTACCAGTGATTTTAGGGGTCAATGACAATGAAGGCTACTATTTTTCAGGCCTAGAGGACTCTTCTACTCTTACGCGTTTTAAATTTGAAATCGGTTTACCAAGAAATTTACGGATTCCAGCTGAACTCAAGAAAGAAGTTTGTCAGAAATTGCACAAGATGTACATGAGCGAGGATTCAAATTCAATGGAGAGTATTTTGAAGTTTTTGAAGTACCATGCTGATCCGTTTTTCTTGTATCCAGCTTTTGCTGAGACAAAACTACTCTTGCAGAATAATGATCTGCCAGTCTATAATTATTATTTTCGATATGATGGATGGAGGAACTTTGTCAAATTGATCTCAGGATTTTGGGCGGAACCAGGGGCGTCCCACGCTGATGATCTTATGTATTTGTTCAGTATTTTTCCAGTACCAACTTTATTTGAGAGTAAAATCATTGACAGAATGACGACGATGTGGACTAATTTCGCTAAATTTGGAGATCCAACACCGGAGAAGACAGACCTTTTACCCATAAAATGGTATCCGACGAATCAAAGTTTTACGAAAACTCTCGTTATCGATAAGGAATTTACAATTACATCATTCAAGCACAACTCATTTGATTTCTGGCACGATCTTTATAAAAAATATCGTAGAAAAAATTAA
Protein
MCVSIKWVVLWSLWAGRLVRQPTPAVRLSNHRLVSGQVAPDGSHVQYFGIPYATVRRRHRFKAAGLEPEWDGVFNAVNEHVRCPQKFADNFVWGQEDCLIINIFHPLYSEVDKLLPVKVFIHGGGFREGSSSRLLFGPDYLVRKNIVLVTFNYRVAVTGFLCLGIKDAPGNAGLKDQVAALKWVQRNIKVFGGDPDNVTIFGESAGAASVSLHIVSPASKGLFHKAILQSGSSISPWAVEFEPIKYASRVAEVLGYKTMDPYQLYDIHMNHTIESIVGAPVPLLESQVAAAQLINSPCIEKSHPGVEPFLLESPFDIFTRGEYNKVPVILGVNDNEGYYFSGLEDSSTLTRFKFEIGLPRNLRIPAELKKEVCQKLHKMYMSEDSNSMESILKFLKYHADPFFLYPAFAETKLLLQNNDLPVYNYYFRYDGWRNFVKLISGFWAEPGASHADDLMYLFSIFPVPTLFESKIIDRMTTMWTNFAKFGDPTPEKTDLLPIKWYPTNQSFTKTLVIDKEFTITSFKHNSFDFWHDLYKKYRRKN
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
M1RMH6
A0A194PP48
A0A194QY52
A0A0B4RYC5
A0A0K0XRM7
A0A2H1VEU9
+ More
A0A2W1BSB9 A0A2H1VDR7 A0A2A4J1Z4 A0A291P0Y6 D5G3G3 E5KG16 A0A2H1WQE3 H9J3R0 A0A0K8TV95 D3GDM8 E5KG15 A0A2S0D875 A0A2A4J7T3 A0A2U3T8K5 A0A1U9X1W6 A0A0F7QIF1 A0A076FR95 H9J3W6 A0A0Y0C087 D3GDN1 A0A2H1WQD3 E5KG19 Q5YJK2 A0A2W1BUD0 A0A1L8D6X8 A0A2W1BXK5 A0A1J0M1W7 A0A0C5C590 A0A194PNF5 A0A1U9X1T0 A0A1U9X1Y4 A0A1U9X1U5 A0A0Y0BFP9 A0A1L8D6K4 A0A0L7LDS4 A0A1U9X1Y2 A0A194R3H8 A0A0F7QIF3 A0A212EJY6 A0A291P0T0 A0A0H4SHU3 A0A2W1BYR0 A0A0K8TVU4 A0A0K8TVW9 A0A1U9X1Y5 A0A0K8TUM8 A0A146JVQ5 A0A1U9X1S7 A0A2H1WRR7 A0A1J0M1C0 A0A2A4J4R6 A4UA25 A0A076FRK9 A0A0K0XRT4 A0A2U3T8J9 A0A2U3T8J4 Q86MC7 A0A0L7L9I4 A0A0F7QJY8 D3GDM9 E5KG14 A0A0C5C1Q4 D2KWC0 A0A194QZJ3 G1C2I3 A0A2W1BSD0 A0A2U3T8K6 A0A076FRM7 A0A0F7QJX1 A0A0K0XRS9 A0A0B4RXS6 B3GS56 A0A2A4J7L0 H9JT66 B1Q140 H9JT67 A0A2A4J6K4 A0A0K8TUN6 A0A2A4J7A4 A0A291P0T5 D2KWB9 A0A0L7L3B6 A0A0K0XRW7 A0A2H1X3E8 A0A0C4JZC5 A0A2W1BV54 A0A0B4RYB9 A0A194PP41 A0A212F4S9 A0A2W1BTK4 A0A2W1BSA9 D3GDL9
A0A2W1BSB9 A0A2H1VDR7 A0A2A4J1Z4 A0A291P0Y6 D5G3G3 E5KG16 A0A2H1WQE3 H9J3R0 A0A0K8TV95 D3GDM8 E5KG15 A0A2S0D875 A0A2A4J7T3 A0A2U3T8K5 A0A1U9X1W6 A0A0F7QIF1 A0A076FR95 H9J3W6 A0A0Y0C087 D3GDN1 A0A2H1WQD3 E5KG19 Q5YJK2 A0A2W1BUD0 A0A1L8D6X8 A0A2W1BXK5 A0A1J0M1W7 A0A0C5C590 A0A194PNF5 A0A1U9X1T0 A0A1U9X1Y4 A0A1U9X1U5 A0A0Y0BFP9 A0A1L8D6K4 A0A0L7LDS4 A0A1U9X1Y2 A0A194R3H8 A0A0F7QIF3 A0A212EJY6 A0A291P0T0 A0A0H4SHU3 A0A2W1BYR0 A0A0K8TVU4 A0A0K8TVW9 A0A1U9X1Y5 A0A0K8TUM8 A0A146JVQ5 A0A1U9X1S7 A0A2H1WRR7 A0A1J0M1C0 A0A2A4J4R6 A4UA25 A0A076FRK9 A0A0K0XRT4 A0A2U3T8J9 A0A2U3T8J4 Q86MC7 A0A0L7L9I4 A0A0F7QJY8 D3GDM9 E5KG14 A0A0C5C1Q4 D2KWC0 A0A194QZJ3 G1C2I3 A0A2W1BSD0 A0A2U3T8K6 A0A076FRM7 A0A0F7QJX1 A0A0K0XRS9 A0A0B4RXS6 B3GS56 A0A2A4J7L0 H9JT66 B1Q140 H9JT67 A0A2A4J6K4 A0A0K8TUN6 A0A2A4J7A4 A0A291P0T5 D2KWB9 A0A0L7L3B6 A0A0K0XRW7 A0A2H1X3E8 A0A0C4JZC5 A0A2W1BV54 A0A0B4RYB9 A0A194PP41 A0A212F4S9 A0A2W1BTK4 A0A2W1BSA9 D3GDL9
EC Number
3.1.1.-
Pubmed
EMBL
KC200269
AGG20204.1
KQ459598
KPI94519.1
KQ460949
KPJ10397.1
+ More
KJ023204 AIY69032.1 KP938888 AKS40369.1 ODYU01001984 SOQ38932.1 KZ149915 PZC77958.1 ODYU01001983 SOQ38931.1 NWSH01003910 PCG65698.1 MF687635 ATJ44561.1 FJ997327 ADF43492.1 HQ116559 ADR64700.1 ODYU01010289 SOQ55289.1 BABH01028961 BABH01028962 BABH01028963 GCVX01000044 JAI18186.1 FJ652460 ACV60244.1 HQ116558 ADR64699.1 KX015862 ARM65391.1 NWSH01002809 PCG67500.1 KX015859 ARM65388.1 KY021868 AQY62757.1 LC017769 BAR64783.1 KF960790 AII21992.1 BABH01028964 KU215372 AMB19666.1 FJ652463 ACV60247.1 SOQ55290.1 HQ116562 ADR64703.1 AY390258 AAR26516.1 PZC77961.1 GEYN01000039 JAV02090.1 PZC77957.1 KU375436 APD13878.1 KP001155 AJN91200.1 KPI94518.1 KY021827 AQY62716.1 KY021871 AQY62760.1 KY021837 AQY62726.1 KU215365 AMB19659.1 GEYN01000040 JAV02089.1 JTDY01001530 KOB73632.1 KY021877 AQY62766.1 KPJ10396.1 LC017774 BAR64788.1 AGBW02014361 OWR41813.1 MF687562 ATJ44488.1 KP677287 AKP92855.1 PZC77960.1 GCVX01000045 JAI18185.1 GCVX01000020 JAI18210.1 KY021875 AQY62764.1 GCVX01000043 JAI18187.1 GEDO01000023 JAP88603.1 KY021828 AQY62717.1 ODYU01010555 SOQ55751.1 KU375425 APD13867.1 NWSH01003373 PCG66430.1 DQ680828 ABH01081.1 KF960776 AII21978.1 KP938884 AKS40365.1 KX015850 ARM65379.1 KX015843 ARM65372.1 AY091503 AAM14415.1 JTDY01002125 KOB72049.1 LC017773 BAR64787.1 FJ652461 ACV60245.1 HQ116557 ADR64698.1 KP001154 AJN91199.1 AB497075 BAI66481.1 KPJ10395.1 JF728802 AEJ38204.1 KZ149953 PZC76535.1 KX015860 ARM65389.1 KF960788 AII21990.1 LC017768 BAR64782.1 KP938879 AKS40360.1 KJ023217 AIY69045.1 EU727142 ACE62801.1 NWSH01002873 PCG67402.1 BABH01034844 EU523535 ACB12414.1 PCG67499.1 GCVX01000026 JAI18204.1 PCG67404.1 MF687559 ATJ44485.1 AB497074 AB299283 BAI66480.1 BAI94515.1 JTDY01003306 KOB69769.1 KP938885 AKS40366.1 ODYU01013095 SOQ59778.1 KC832922 AHF45923.1 PZC76536.1 KJ023199 AIY69027.1 KPI94514.1 AGBW02010320 OWR48730.1 PZC76537.1 PZC76534.1 FJ652451 ACV60235.1
KJ023204 AIY69032.1 KP938888 AKS40369.1 ODYU01001984 SOQ38932.1 KZ149915 PZC77958.1 ODYU01001983 SOQ38931.1 NWSH01003910 PCG65698.1 MF687635 ATJ44561.1 FJ997327 ADF43492.1 HQ116559 ADR64700.1 ODYU01010289 SOQ55289.1 BABH01028961 BABH01028962 BABH01028963 GCVX01000044 JAI18186.1 FJ652460 ACV60244.1 HQ116558 ADR64699.1 KX015862 ARM65391.1 NWSH01002809 PCG67500.1 KX015859 ARM65388.1 KY021868 AQY62757.1 LC017769 BAR64783.1 KF960790 AII21992.1 BABH01028964 KU215372 AMB19666.1 FJ652463 ACV60247.1 SOQ55290.1 HQ116562 ADR64703.1 AY390258 AAR26516.1 PZC77961.1 GEYN01000039 JAV02090.1 PZC77957.1 KU375436 APD13878.1 KP001155 AJN91200.1 KPI94518.1 KY021827 AQY62716.1 KY021871 AQY62760.1 KY021837 AQY62726.1 KU215365 AMB19659.1 GEYN01000040 JAV02089.1 JTDY01001530 KOB73632.1 KY021877 AQY62766.1 KPJ10396.1 LC017774 BAR64788.1 AGBW02014361 OWR41813.1 MF687562 ATJ44488.1 KP677287 AKP92855.1 PZC77960.1 GCVX01000045 JAI18185.1 GCVX01000020 JAI18210.1 KY021875 AQY62764.1 GCVX01000043 JAI18187.1 GEDO01000023 JAP88603.1 KY021828 AQY62717.1 ODYU01010555 SOQ55751.1 KU375425 APD13867.1 NWSH01003373 PCG66430.1 DQ680828 ABH01081.1 KF960776 AII21978.1 KP938884 AKS40365.1 KX015850 ARM65379.1 KX015843 ARM65372.1 AY091503 AAM14415.1 JTDY01002125 KOB72049.1 LC017773 BAR64787.1 FJ652461 ACV60245.1 HQ116557 ADR64698.1 KP001154 AJN91199.1 AB497075 BAI66481.1 KPJ10395.1 JF728802 AEJ38204.1 KZ149953 PZC76535.1 KX015860 ARM65389.1 KF960788 AII21990.1 LC017768 BAR64782.1 KP938879 AKS40360.1 KJ023217 AIY69045.1 EU727142 ACE62801.1 NWSH01002873 PCG67402.1 BABH01034844 EU523535 ACB12414.1 PCG67499.1 GCVX01000026 JAI18204.1 PCG67404.1 MF687559 ATJ44485.1 AB497074 AB299283 BAI66480.1 BAI94515.1 JTDY01003306 KOB69769.1 KP938885 AKS40366.1 ODYU01013095 SOQ59778.1 KC832922 AHF45923.1 PZC76536.1 KJ023199 AIY69027.1 KPI94514.1 AGBW02010320 OWR48730.1 PZC76537.1 PZC76534.1 FJ652451 ACV60235.1
Proteomes
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
M1RMH6
A0A194PP48
A0A194QY52
A0A0B4RYC5
A0A0K0XRM7
A0A2H1VEU9
+ More
A0A2W1BSB9 A0A2H1VDR7 A0A2A4J1Z4 A0A291P0Y6 D5G3G3 E5KG16 A0A2H1WQE3 H9J3R0 A0A0K8TV95 D3GDM8 E5KG15 A0A2S0D875 A0A2A4J7T3 A0A2U3T8K5 A0A1U9X1W6 A0A0F7QIF1 A0A076FR95 H9J3W6 A0A0Y0C087 D3GDN1 A0A2H1WQD3 E5KG19 Q5YJK2 A0A2W1BUD0 A0A1L8D6X8 A0A2W1BXK5 A0A1J0M1W7 A0A0C5C590 A0A194PNF5 A0A1U9X1T0 A0A1U9X1Y4 A0A1U9X1U5 A0A0Y0BFP9 A0A1L8D6K4 A0A0L7LDS4 A0A1U9X1Y2 A0A194R3H8 A0A0F7QIF3 A0A212EJY6 A0A291P0T0 A0A0H4SHU3 A0A2W1BYR0 A0A0K8TVU4 A0A0K8TVW9 A0A1U9X1Y5 A0A0K8TUM8 A0A146JVQ5 A0A1U9X1S7 A0A2H1WRR7 A0A1J0M1C0 A0A2A4J4R6 A4UA25 A0A076FRK9 A0A0K0XRT4 A0A2U3T8J9 A0A2U3T8J4 Q86MC7 A0A0L7L9I4 A0A0F7QJY8 D3GDM9 E5KG14 A0A0C5C1Q4 D2KWC0 A0A194QZJ3 G1C2I3 A0A2W1BSD0 A0A2U3T8K6 A0A076FRM7 A0A0F7QJX1 A0A0K0XRS9 A0A0B4RXS6 B3GS56 A0A2A4J7L0 H9JT66 B1Q140 H9JT67 A0A2A4J6K4 A0A0K8TUN6 A0A2A4J7A4 A0A291P0T5 D2KWB9 A0A0L7L3B6 A0A0K0XRW7 A0A2H1X3E8 A0A0C4JZC5 A0A2W1BV54 A0A0B4RYB9 A0A194PP41 A0A212F4S9 A0A2W1BTK4 A0A2W1BSA9 D3GDL9
A0A2W1BSB9 A0A2H1VDR7 A0A2A4J1Z4 A0A291P0Y6 D5G3G3 E5KG16 A0A2H1WQE3 H9J3R0 A0A0K8TV95 D3GDM8 E5KG15 A0A2S0D875 A0A2A4J7T3 A0A2U3T8K5 A0A1U9X1W6 A0A0F7QIF1 A0A076FR95 H9J3W6 A0A0Y0C087 D3GDN1 A0A2H1WQD3 E5KG19 Q5YJK2 A0A2W1BUD0 A0A1L8D6X8 A0A2W1BXK5 A0A1J0M1W7 A0A0C5C590 A0A194PNF5 A0A1U9X1T0 A0A1U9X1Y4 A0A1U9X1U5 A0A0Y0BFP9 A0A1L8D6K4 A0A0L7LDS4 A0A1U9X1Y2 A0A194R3H8 A0A0F7QIF3 A0A212EJY6 A0A291P0T0 A0A0H4SHU3 A0A2W1BYR0 A0A0K8TVU4 A0A0K8TVW9 A0A1U9X1Y5 A0A0K8TUM8 A0A146JVQ5 A0A1U9X1S7 A0A2H1WRR7 A0A1J0M1C0 A0A2A4J4R6 A4UA25 A0A076FRK9 A0A0K0XRT4 A0A2U3T8J9 A0A2U3T8J4 Q86MC7 A0A0L7L9I4 A0A0F7QJY8 D3GDM9 E5KG14 A0A0C5C1Q4 D2KWC0 A0A194QZJ3 G1C2I3 A0A2W1BSD0 A0A2U3T8K6 A0A076FRM7 A0A0F7QJX1 A0A0K0XRS9 A0A0B4RXS6 B3GS56 A0A2A4J7L0 H9JT66 B1Q140 H9JT67 A0A2A4J6K4 A0A0K8TUN6 A0A2A4J7A4 A0A291P0T5 D2KWB9 A0A0L7L3B6 A0A0K0XRW7 A0A2H1X3E8 A0A0C4JZC5 A0A2W1BV54 A0A0B4RYB9 A0A194PP41 A0A212F4S9 A0A2W1BTK4 A0A2W1BSA9 D3GDL9
PDB
5IVK
E-value=1.17565e-57,
Score=567
Ontologies
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.537510
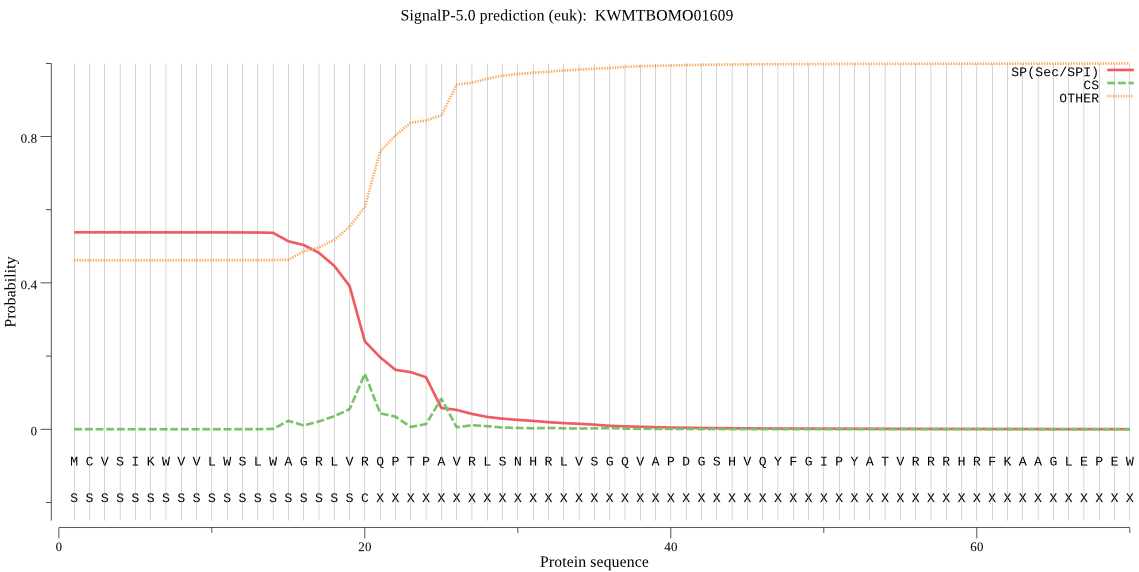
Length:
541
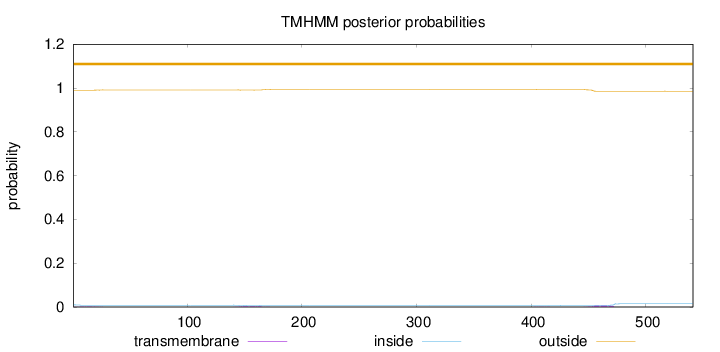
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23038
Exp number, first 60 AAs:
0.02606
Total prob of N-in:
0.00989
outside
1 - 541
Population Genetic Test Statistics
Pi
217.300328
Theta
208.523828
Tajima's D
0.012169
CLR
0.437133
CSRT
0.369881505924704
Interpretation
Uncertain
