Gene
KWMTBOMO01606 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006085
Annotation
PREDICTED:_nucleolar_protein_58_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.974 Nuclear Reliability : 1.59
Sequence
CDS
ATGTTGGTTCTATTCGAGACACCGGCGGGCTATGCCATATTTAAGTTGCTGGATGAGTCTAAGCTGAGTGAGATTGACAACTTGTACCAAGAATTTAACACTCCAGAAGGGGCTTCCACAGTGGTTAAGTTAAAGAACTTTGTAAAGTTTGAGGACACAACAGAAGCTTTGGCAGCCACTACTGCAGCTATTGAGGGCAAGATATCTAAGCCCCTAAAGAAAGCTCTTAAGAAGTATGTTTGTAAAGAGGTTCAGGATCAACTCCTTGTCGGTGACTCTAAGCTGGGAAGTGCCATTAGAGAAAAATTTGACTTACAATGTGTATCAAATACTAATGTCCAGGAACTGCTCCGGTGTATTCGCTCACAGATGGACAGTCTCCTAGCTGGACTTCCTAAAAAAGAAATGACAGCTATGGCTTTGGGCCTGGCTCACTCTCTTTCGAGGTACAAATTAAAGTTCTCTCCTGACAAAATAGACACTATGATTGTTCAAGCTCAGTGTTTGCTTGATGATCTTGACAAGGAATTAAACAACTACATCATGAGGTGCCGTGAATGGTATGGTTGGCATTTCCCAGAGCTTGGGAAAATCATCACTGATAACACTTTGTTTGTAAAAGTGGTAAAACTCATTGGCACACGAGACTATGCATCCAAGACTGATTTATCTGACATTTTACCTGAAGACTTAGAAGAGAAAGTTAAGGAAGCAGCTGAGATCTCCATGGGAACTGAAATATCTGAAGACGACATTATGAACATACAGAATCTGTGTGATGAGATTATTTCCATCACAGACTACAGGACTCATTTAACAGACTACTTGAAGGCGCGTATGATGGCAATGGCACCAAACCTCACTGTACTCATTGGTGAACACATTGGAGCACGACTTATTGCTCATGCAGGCTCACTAATGAATTTGGCAAAACATCCTGCTTCTACAATACAGATTTTTGGTGCAGAAAAAGCTCTTTTCCGTGCCTTAAAAACGAAGAAAGACACACCAAAGTATGGATTGATATATCACGCTCAGTTAGTGGGCCAGTGCAGTACAAAAAACAAAGGTAAAATGTCAAGAATGTTGGCTGCTAAGGCAGCACTTGCGACTAGAGTTGATGCATTTGGAGAAGATGTGTCTTTTGAGCTCGGAGCTGAACATAAAGTGAAATTAGAGAACCGCCTGCGACTACTGGAAGAAGGAAACCTGCGTAGGATTAGTGGAACAGGGAAAGCAAAAATCAAATTTGAGAAATATCACAGTAAAAGTGAAGTCTATCAATATCCTGATGCAAGTGATAACACAATTGACAAGAAGCCGATCAAGCGAGAACACTCTCCAGATGAAGACCAAGTGTCAGCCAAAAAGATAAAACTAGAGAATGATGTGTCACTTAAAAATATAAAAACTGAAAACGAAGAACCGGCAGAAGACGCGCTCCCAGAACCCAGCTCTGAAAAGAAGAAAAAGAAGAAACGGAAATCAGAAGTAAAAACTGAACCAGAACCAGAACCCGAGGTCTCACTTGAAGAACCCAAGAGTGAAAAAAAGAAGAAAAAGAAGCGGCAGTCTCAGGCCGATGAATAG
Protein
MLVLFETPAGYAIFKLLDESKLSEIDNLYQEFNTPEGASTVVKLKNFVKFEDTTEALAATTAAIEGKISKPLKKALKKYVCKEVQDQLLVGDSKLGSAIREKFDLQCVSNTNVQELLRCIRSQMDSLLAGLPKKEMTAMALGLAHSLSRYKLKFSPDKIDTMIVQAQCLLDDLDKELNNYIMRCREWYGWHFPELGKIITDNTLFVKVVKLIGTRDYASKTDLSDILPEDLEEKVKEAAEISMGTEISEDDIMNIQNLCDEIISITDYRTHLTDYLKARMMAMAPNLTVLIGEHIGARLIAHAGSLMNLAKHPASTIQIFGAEKALFRALKTKKDTPKYGLIYHAQLVGQCSTKNKGKMSRMLAAKAALATRVDAFGEDVSFELGAEHKVKLENRLRLLEEGNLRRISGTGKAKIKFEKYHSKSEVYQYPDASDNTIDKKPIKREHSPDEDQVSAKKIKLENDVSLKNIKTENEEPAEDALPEPSSEKKKKKKRKSEVKTEPEPEPEVSLEEPKSEKKKKKKRQSQADE
Summary
Uniprot
H9J993
A0A2H1X0C1
A0A2A4JKJ4
A0A2W1BVK5
A0A194Q0C8
A0A194R708
+ More
S4P206 A0A212FGN2 A0A1E1WCL6 A0A0L7LG86 A0A087ZZZ5 A0A2A3E580 A0A154PKI1 B4HXZ8 B4N068 D6WGW9 A0ANZ0 A0A1W4X4X6 A0ANY9 B4Q595 Q9U5W4 A0A1W4UQ94 Q9VM69 A0A0L7R3J5 B3MKR6 B4NZZ2 V5H0A7 B5DK75 B3N662 B4LRP1 A0A1L8DHH6 A0A1L8DH61 B4GKZ4 A0A1L8DHE6 A0A1I8QEY6 A0A1A9V9M1 A0A3B0KD38 A0A1Q3F065 A0A1Y1MP76 D3TND0 A0A067RI98 B0X5E5 A0A0C9QIS7 A0A1A9W031 A0A0M4E4A3 A0A1B0A509 B4JQL6 A0A0L0BQJ7 A0A1A9XGV6 A0A1B0BJH4 A0A336LXS0 B4KJW5 A0A2P8XVX0 A0A0N0U576 A0A1B0FF83 U5EYK2 A0A1I8MP78 A0A1J1I5D9 A0A1B6FBC6 A0A182RFT9 A0A0M9A283 W8BSS4 A0A1B6L2R7 A0A1B6HPN4 K7ILS5 A0A2M4BJ71 A0A2M4BJE9 A0A2M4BJ45 A0A2M4BJ39 A0A0K8UHY0 A0A182FQN3 A0A2M4BJC8 A0A034VYR3 A0A182TDW1 A0A0A1WU25 A0A2M4ADG7 A0A2M4AD97 A0A182VCD0 Q7QK65 A0A0P5P451 A0A182MG25 A0A0N8AK20 A0A182QC14 A0A182LQQ0 A0A0P6GUL0 A0A0P5D0G8 A0A0P4WWJ6 A0A2M3Z4S2 A0A2M3Z4E5 A0A0N8C939 A0A0P5BB74 A0A0P4YCA3 A0A0P4WYQ8 A0A0P5D1V1 A0A0P5QYW7 A0A0P5P4L3 W5J4V6 A0A0P5TSS0 A0A0P5FL43 A0A0P5DS21
S4P206 A0A212FGN2 A0A1E1WCL6 A0A0L7LG86 A0A087ZZZ5 A0A2A3E580 A0A154PKI1 B4HXZ8 B4N068 D6WGW9 A0ANZ0 A0A1W4X4X6 A0ANY9 B4Q595 Q9U5W4 A0A1W4UQ94 Q9VM69 A0A0L7R3J5 B3MKR6 B4NZZ2 V5H0A7 B5DK75 B3N662 B4LRP1 A0A1L8DHH6 A0A1L8DH61 B4GKZ4 A0A1L8DHE6 A0A1I8QEY6 A0A1A9V9M1 A0A3B0KD38 A0A1Q3F065 A0A1Y1MP76 D3TND0 A0A067RI98 B0X5E5 A0A0C9QIS7 A0A1A9W031 A0A0M4E4A3 A0A1B0A509 B4JQL6 A0A0L0BQJ7 A0A1A9XGV6 A0A1B0BJH4 A0A336LXS0 B4KJW5 A0A2P8XVX0 A0A0N0U576 A0A1B0FF83 U5EYK2 A0A1I8MP78 A0A1J1I5D9 A0A1B6FBC6 A0A182RFT9 A0A0M9A283 W8BSS4 A0A1B6L2R7 A0A1B6HPN4 K7ILS5 A0A2M4BJ71 A0A2M4BJE9 A0A2M4BJ45 A0A2M4BJ39 A0A0K8UHY0 A0A182FQN3 A0A2M4BJC8 A0A034VYR3 A0A182TDW1 A0A0A1WU25 A0A2M4ADG7 A0A2M4AD97 A0A182VCD0 Q7QK65 A0A0P5P451 A0A182MG25 A0A0N8AK20 A0A182QC14 A0A182LQQ0 A0A0P6GUL0 A0A0P5D0G8 A0A0P4WWJ6 A0A2M3Z4S2 A0A2M3Z4E5 A0A0N8C939 A0A0P5BB74 A0A0P4YCA3 A0A0P4WYQ8 A0A0P5D1V1 A0A0P5QYW7 A0A0P5P4L3 W5J4V6 A0A0P5TSS0 A0A0P5FL43 A0A0P5DS21
Pubmed
19121390
28756777
26354079
23622113
22118469
26227816
+ More
17994087 18362917 19820115 16951084 22936249 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 28004739 20353571 24845553 26108605 29403074 25315136 24495485 20075255 25348373 25830018 12364791 14747013 17210077 20966253 20920257 23761445
17994087 18362917 19820115 16951084 22936249 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 28004739 20353571 24845553 26108605 29403074 25315136 24495485 20075255 25348373 25830018 12364791 14747013 17210077 20966253 20920257 23761445
EMBL
BABH01004300
ODYU01012473
SOQ58800.1
NWSH01001259
PCG71953.1
KZ149934
+ More
PZC77227.1 KQ459584 KPI98449.1 KQ460644 KPJ13304.1 GAIX01012235 JAA80325.1 AGBW02008633 OWR52895.1 GDQN01006331 JAT84723.1 JTDY01001244 KOB74420.1 KZ288387 PBC26416.1 KQ434946 KZC12356.1 CH480818 EDW51928.1 CH963920 EDW78003.1 KQ971321 EFA00147.1 AM294378 CAL26308.1 AM294377 CAL26307.1 CM000361 CM002910 EDX04028.1 KMY88647.1 AJ249465 AM294373 AM294374 AM294375 AM294376 AM294379 AM294380 FM245446 FM245447 FM245449 FM245450 FM245451 FM245452 FM245454 CAB60723.1 CAL26303.1 CAL26304.1 CAL26305.1 CAL26306.1 CAL26309.1 CAL26310.1 CAR93372.1 CAR93373.1 CAR93375.1 CAR93376.1 CAR93377.1 CAR93378.1 CAR93380.1 AE014134 AY061401 FM245448 FM245453 AAF52455.2 AAL28949.1 CAR93374.1 CAR93379.1 KQ414663 KOC65399.1 CH902620 EDV31597.1 CM000157 EDW87819.1 GALX01002208 JAB66258.1 CH379062 EDY70701.1 CH954177 EDV59149.1 CH940649 EDW64643.1 GFDF01008290 JAV05794.1 GFDF01008323 JAV05761.1 CH479184 EDW37310.1 GFDF01008289 JAV05795.1 OUUW01000006 SPP81558.1 GFDL01014090 JAV20955.1 GEZM01025389 JAV87479.1 EZ422932 ADD19208.1 KK852458 KDR23571.1 DS232376 EDS40830.1 GBYB01003434 JAG73201.1 CP012523 ALC38646.1 CH916372 EDV99196.1 JRES01001511 KNC22355.1 JXJN01015395 UFQT01000283 SSX22764.1 CH933807 EDW12568.1 PYGN01001265 PSN36152.1 KQ435789 KOX74452.1 CCAG010014241 GANO01000304 JAB59567.1 CVRI01000040 CRK94804.1 GECZ01022262 JAS47507.1 KOX74453.1 GAMC01004349 GAMC01004348 JAC02208.1 GEBQ01022046 JAT17931.1 GECU01031058 JAS76648.1 GGFJ01003941 MBW53082.1 GGFJ01003940 MBW53081.1 GGFJ01003939 MBW53080.1 GGFJ01003945 MBW53086.1 GDHF01026148 JAI26166.1 GGFJ01003942 MBW53083.1 GAKP01011922 GAKP01011920 JAC47030.1 GBXI01011980 JAD02312.1 GGFK01005441 MBW38762.1 GGFK01005428 MBW38749.1 AAAB01008799 EAA03754.5 GDIQ01138290 JAL13436.1 AXCM01003899 GDIQ01268099 JAJ83625.1 AXCN02001550 GDIQ01041325 JAN53412.1 GDIP01164574 JAJ58828.1 GDIP01251532 JAI71869.1 GGFM01002766 MBW23517.1 GGFM01002641 MBW23392.1 GDIP01251531 GDIQ01177213 GDIQ01175260 GDIQ01102747 LRGB01002384 JAI71870.1 JAL48979.1 KZS07929.1 GDIP01191232 GDIP01092294 JAJ32170.1 GDIP01229531 JAI93870.1 GDIP01251530 JAI71871.1 GDIP01167616 JAJ55786.1 GDIQ01110342 JAL41384.1 GDIQ01133789 JAL17937.1 ADMH02002134 ETN58428.1 GDIP01122086 JAL81628.1 GDIQ01252956 JAJ98768.1 GDIP01158327 JAJ65075.1
PZC77227.1 KQ459584 KPI98449.1 KQ460644 KPJ13304.1 GAIX01012235 JAA80325.1 AGBW02008633 OWR52895.1 GDQN01006331 JAT84723.1 JTDY01001244 KOB74420.1 KZ288387 PBC26416.1 KQ434946 KZC12356.1 CH480818 EDW51928.1 CH963920 EDW78003.1 KQ971321 EFA00147.1 AM294378 CAL26308.1 AM294377 CAL26307.1 CM000361 CM002910 EDX04028.1 KMY88647.1 AJ249465 AM294373 AM294374 AM294375 AM294376 AM294379 AM294380 FM245446 FM245447 FM245449 FM245450 FM245451 FM245452 FM245454 CAB60723.1 CAL26303.1 CAL26304.1 CAL26305.1 CAL26306.1 CAL26309.1 CAL26310.1 CAR93372.1 CAR93373.1 CAR93375.1 CAR93376.1 CAR93377.1 CAR93378.1 CAR93380.1 AE014134 AY061401 FM245448 FM245453 AAF52455.2 AAL28949.1 CAR93374.1 CAR93379.1 KQ414663 KOC65399.1 CH902620 EDV31597.1 CM000157 EDW87819.1 GALX01002208 JAB66258.1 CH379062 EDY70701.1 CH954177 EDV59149.1 CH940649 EDW64643.1 GFDF01008290 JAV05794.1 GFDF01008323 JAV05761.1 CH479184 EDW37310.1 GFDF01008289 JAV05795.1 OUUW01000006 SPP81558.1 GFDL01014090 JAV20955.1 GEZM01025389 JAV87479.1 EZ422932 ADD19208.1 KK852458 KDR23571.1 DS232376 EDS40830.1 GBYB01003434 JAG73201.1 CP012523 ALC38646.1 CH916372 EDV99196.1 JRES01001511 KNC22355.1 JXJN01015395 UFQT01000283 SSX22764.1 CH933807 EDW12568.1 PYGN01001265 PSN36152.1 KQ435789 KOX74452.1 CCAG010014241 GANO01000304 JAB59567.1 CVRI01000040 CRK94804.1 GECZ01022262 JAS47507.1 KOX74453.1 GAMC01004349 GAMC01004348 JAC02208.1 GEBQ01022046 JAT17931.1 GECU01031058 JAS76648.1 GGFJ01003941 MBW53082.1 GGFJ01003940 MBW53081.1 GGFJ01003939 MBW53080.1 GGFJ01003945 MBW53086.1 GDHF01026148 JAI26166.1 GGFJ01003942 MBW53083.1 GAKP01011922 GAKP01011920 JAC47030.1 GBXI01011980 JAD02312.1 GGFK01005441 MBW38762.1 GGFK01005428 MBW38749.1 AAAB01008799 EAA03754.5 GDIQ01138290 JAL13436.1 AXCM01003899 GDIQ01268099 JAJ83625.1 AXCN02001550 GDIQ01041325 JAN53412.1 GDIP01164574 JAJ58828.1 GDIP01251532 JAI71869.1 GGFM01002766 MBW23517.1 GGFM01002641 MBW23392.1 GDIP01251531 GDIQ01177213 GDIQ01175260 GDIQ01102747 LRGB01002384 JAI71870.1 JAL48979.1 KZS07929.1 GDIP01191232 GDIP01092294 JAJ32170.1 GDIP01229531 JAI93870.1 GDIP01251530 JAI71871.1 GDIP01167616 JAJ55786.1 GDIQ01110342 JAL41384.1 GDIQ01133789 JAL17937.1 ADMH02002134 ETN58428.1 GDIP01122086 JAL81628.1 GDIQ01252956 JAJ98768.1 GDIP01158327 JAJ65075.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000005203 UP000242457 UP000076502 UP000001292 UP000007798 UP000007266 UP000192223 UP000000304 UP000192221 UP000000803 UP000053825 UP000007801 UP000002282 UP000001819 UP000008711 UP000008792 UP000008744 UP000095300 UP000078200 UP000268350 UP000027135 UP000002320 UP000091820 UP000092553 UP000092445 UP000001070 UP000037069 UP000092443 UP000092460 UP000009192 UP000245037 UP000053105 UP000092444 UP000095301 UP000183832 UP000075900 UP000002358 UP000069272 UP000075902 UP000075903 UP000007062 UP000075883 UP000075886 UP000075882 UP000076858 UP000000673
UP000005203 UP000242457 UP000076502 UP000001292 UP000007798 UP000007266 UP000192223 UP000000304 UP000192221 UP000000803 UP000053825 UP000007801 UP000002282 UP000001819 UP000008711 UP000008792 UP000008744 UP000095300 UP000078200 UP000268350 UP000027135 UP000002320 UP000091820 UP000092553 UP000092445 UP000001070 UP000037069 UP000092443 UP000092460 UP000009192 UP000245037 UP000053105 UP000092444 UP000095301 UP000183832 UP000075900 UP000002358 UP000069272 UP000075902 UP000075903 UP000007062 UP000075883 UP000075886 UP000075882 UP000076858 UP000000673
Interpro
IPR029012
Helix_hairpin_bin_sf
+ More
IPR012976 NOSIC
IPR036070 Nop_dom_sf
IPR002687 Nop_dom
IPR012974 NOP5_N
IPR042239 Nop_C
IPR027417 P-loop_NTPase
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR007694 DNA_helicase_DnaB-like_C
IPR003593 AAA+_ATPase
IPR035979 RBD_domain_sf
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR016189 Transl_init_fac_IF2/IF5_N
IPR002735 Transl_init_fac_IF2/IF5
IPR012976 NOSIC
IPR036070 Nop_dom_sf
IPR002687 Nop_dom
IPR012974 NOP5_N
IPR042239 Nop_C
IPR027417 P-loop_NTPase
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR007694 DNA_helicase_DnaB-like_C
IPR003593 AAA+_ATPase
IPR035979 RBD_domain_sf
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR016189 Transl_init_fac_IF2/IF5_N
IPR002735 Transl_init_fac_IF2/IF5
SUPFAM
Gene 3D
ProteinModelPortal
H9J993
A0A2H1X0C1
A0A2A4JKJ4
A0A2W1BVK5
A0A194Q0C8
A0A194R708
+ More
S4P206 A0A212FGN2 A0A1E1WCL6 A0A0L7LG86 A0A087ZZZ5 A0A2A3E580 A0A154PKI1 B4HXZ8 B4N068 D6WGW9 A0ANZ0 A0A1W4X4X6 A0ANY9 B4Q595 Q9U5W4 A0A1W4UQ94 Q9VM69 A0A0L7R3J5 B3MKR6 B4NZZ2 V5H0A7 B5DK75 B3N662 B4LRP1 A0A1L8DHH6 A0A1L8DH61 B4GKZ4 A0A1L8DHE6 A0A1I8QEY6 A0A1A9V9M1 A0A3B0KD38 A0A1Q3F065 A0A1Y1MP76 D3TND0 A0A067RI98 B0X5E5 A0A0C9QIS7 A0A1A9W031 A0A0M4E4A3 A0A1B0A509 B4JQL6 A0A0L0BQJ7 A0A1A9XGV6 A0A1B0BJH4 A0A336LXS0 B4KJW5 A0A2P8XVX0 A0A0N0U576 A0A1B0FF83 U5EYK2 A0A1I8MP78 A0A1J1I5D9 A0A1B6FBC6 A0A182RFT9 A0A0M9A283 W8BSS4 A0A1B6L2R7 A0A1B6HPN4 K7ILS5 A0A2M4BJ71 A0A2M4BJE9 A0A2M4BJ45 A0A2M4BJ39 A0A0K8UHY0 A0A182FQN3 A0A2M4BJC8 A0A034VYR3 A0A182TDW1 A0A0A1WU25 A0A2M4ADG7 A0A2M4AD97 A0A182VCD0 Q7QK65 A0A0P5P451 A0A182MG25 A0A0N8AK20 A0A182QC14 A0A182LQQ0 A0A0P6GUL0 A0A0P5D0G8 A0A0P4WWJ6 A0A2M3Z4S2 A0A2M3Z4E5 A0A0N8C939 A0A0P5BB74 A0A0P4YCA3 A0A0P4WYQ8 A0A0P5D1V1 A0A0P5QYW7 A0A0P5P4L3 W5J4V6 A0A0P5TSS0 A0A0P5FL43 A0A0P5DS21
S4P206 A0A212FGN2 A0A1E1WCL6 A0A0L7LG86 A0A087ZZZ5 A0A2A3E580 A0A154PKI1 B4HXZ8 B4N068 D6WGW9 A0ANZ0 A0A1W4X4X6 A0ANY9 B4Q595 Q9U5W4 A0A1W4UQ94 Q9VM69 A0A0L7R3J5 B3MKR6 B4NZZ2 V5H0A7 B5DK75 B3N662 B4LRP1 A0A1L8DHH6 A0A1L8DH61 B4GKZ4 A0A1L8DHE6 A0A1I8QEY6 A0A1A9V9M1 A0A3B0KD38 A0A1Q3F065 A0A1Y1MP76 D3TND0 A0A067RI98 B0X5E5 A0A0C9QIS7 A0A1A9W031 A0A0M4E4A3 A0A1B0A509 B4JQL6 A0A0L0BQJ7 A0A1A9XGV6 A0A1B0BJH4 A0A336LXS0 B4KJW5 A0A2P8XVX0 A0A0N0U576 A0A1B0FF83 U5EYK2 A0A1I8MP78 A0A1J1I5D9 A0A1B6FBC6 A0A182RFT9 A0A0M9A283 W8BSS4 A0A1B6L2R7 A0A1B6HPN4 K7ILS5 A0A2M4BJ71 A0A2M4BJE9 A0A2M4BJ45 A0A2M4BJ39 A0A0K8UHY0 A0A182FQN3 A0A2M4BJC8 A0A034VYR3 A0A182TDW1 A0A0A1WU25 A0A2M4ADG7 A0A2M4AD97 A0A182VCD0 Q7QK65 A0A0P5P451 A0A182MG25 A0A0N8AK20 A0A182QC14 A0A182LQQ0 A0A0P6GUL0 A0A0P5D0G8 A0A0P4WWJ6 A0A2M3Z4S2 A0A2M3Z4E5 A0A0N8C939 A0A0P5BB74 A0A0P4YCA3 A0A0P4WYQ8 A0A0P5D1V1 A0A0P5QYW7 A0A0P5P4L3 W5J4V6 A0A0P5TSS0 A0A0P5FL43 A0A0P5DS21
PDB
5WYK
E-value=6.19427e-98,
Score=914
Ontologies
GO
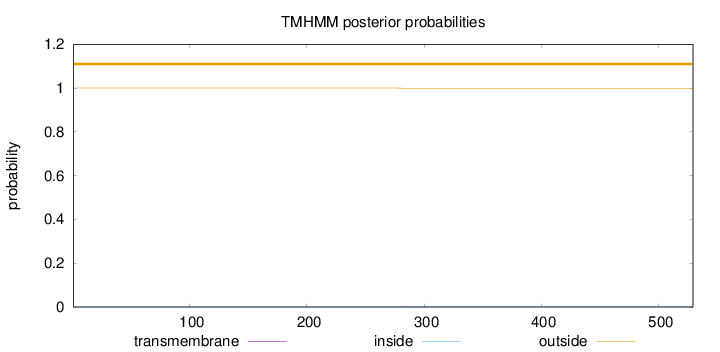
Topology
Length:
529
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.017
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00033
outside
1 - 529
Population Genetic Test Statistics
Pi
16.471589
Theta
22.52322
Tajima's D
0.454087
CLR
0.164961
CSRT
0.512374381280936
Interpretation
Uncertain
