Gene
KWMTBOMO01604 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014304
Annotation
alkaline_phosphatase_[Bombyx_mandarina]
Full name
Alkaline phosphatase
+ More
Membrane-bound alkaline phosphatase
Membrane-bound alkaline phosphatase
Location in the cell
PlasmaMembrane Reliability : 2.634
Sequence
CDS
ATGGTGCGTTTGTTGACACTCCTCTCGCTGCTCGCCGCCGGCTCCTGCGGCCGTCGATATGGATTAGATAAGGACGGTTACCACCGGGATGACGTGGGGTCGAGGCGCTCCCTGCAGACGCCCTCAACGCCGGCCCCCGAGCTGGAGTCGGAGTACTGGTCGCGCGACGCCCAGAGTGAGCTGGGGGAGCGCGCCTGGTACGACGGCAGCAGCGGGTACGCGCGGAACGTCGTCATGTTCCTCGGAGACGGCATGTCCGTGGCCACACTCACGGCCGCCCGCACGCTGCTCGGCCAGCGCCGGGGACAGACTGGAGAGGAGTCCCGATTGTCTTTCGAACATTTTCCAACTGTCGGACTTTCTAAGACCTACTGCCTAGACGCCCAGGTAGCGGACTCGGCTTGCTCGGCCTCCGCCTACCTGTGCGGTGCAAAGGCTAACCTGGGCACCATCGGTGTGTCGGGACACGTGGCGCGACACCACTGTACTGCGGCCACCGACGCTGCACACCAGCTCGCCTCCATCGCCTCCTGGGCTCTCGACGCCGACAGGGACGCCGGGATCGTGACCACCACCCGCGTCACGCACGCCTCGCCGGCCGGAGCGTACGCCCACACCGCCGACCGGAACTGGGAGAGCGACGGTGACGTTACCGCCGGCTGCAGCGGCCACGCTCAGCTGGACATCGCGCAGCAACTCGTCCACGCGCGTCCCGGGAAAGACTTCAAGGTTATTTTGGGAGGCGGAAGAAGAGAGTTCCTACCCAACACTATGACGGACGAAGAGGGCAGTAGAGGAAGAAGATACGATGGGCGAAATCTGATCTCAGAGTGGCTCACTGAGAAGGACAGTCGCGGCGTCACGCACGAATACGTGTGGAACAGACAACAGCTGATGGAGGTCTCCGAAGAGTTGCCAGAGTACCTGCTGGGACTATTCGAACCGAGTCACTTACAATATCATATGGAGGCAAACCATACCATGGAGCCGACCCTGGCCGAGCTCACGGAAGTGGCGATCAAATCGTTGAGTCGCAACAAGGAAGGATTCTTCTTGTTTGTCGAGGGAGGCAGGATCGACCACGCGCACCACGAGAACTTGGCCCACCTCGCTCTCGACGAGACCATCGAACTAAGCGCGGCGGTGGAGAAAGCACGGGAGCTGCTCTCGGAGCAGGATTCATTAATCGTAGTGACGGCGGACCACGCTCACGTCATGTCCATCAGCGGGTACACACAGCGCGGCGGGGACATCCTCGGCCCCTCGGACGCGCTGGGCGACGACGGCATCCCCTACATGACGCTCTCGTATGGCAACGGTCCCGGGTACCGCGAGCCGAGGGGCGGGCAGCGCGTCGACCCGACACGACAAGATTACCGAGGGTCAGAGTACGTGTACCCGGCCGCAGTGCCGCTGGACTCGGAGACTCACGGCGGAGACGACGTGGCGGTATTCGCCTGGGGGCCTCACCACGCCCTCTTCTCTGGCCTCTACGAGCAGAGTCACATCCCGCATCGCATGGCGTACGCAGCATGCATCGGTCCCGGCAAGCACGCTTGCCAACATTCACCACATTCAACTACTTAA
Protein
MVRLLTLLSLLAAGSCGRRYGLDKDGYHRDDVGSRRSLQTPSTPAPELESEYWSRDAQSELGERAWYDGSSGYARNVVMFLGDGMSVATLTAARTLLGQRRGQTGEESRLSFEHFPTVGLSKTYCLDAQVADSACSASAYLCGAKANLGTIGVSGHVARHHCTAATDAAHQLASIASWALDADRDAGIVTTTRVTHASPAGAYAHTADRNWESDGDVTAGCSGHAQLDIAQQLVHARPGKDFKVILGGGRREFLPNTMTDEEGSRGRRYDGRNLISEWLTEKDSRGVTHEYVWNRQQLMEVSEELPEYLLGLFEPSHLQYHMEANHTMEPTLAELTEVAIKSLSRNKEGFFLFVEGGRIDHAHHENLAHLALDETIELSAAVEKARELLSEQDSLIVVTADHAHVMSISGYTQRGGDILGPSDALGDDGIPYMTLSYGNGPGYREPRGGQRVDPTRQDYRGSEYVYPAAVPLDSETHGGDDVAVFAWGPHHALFSGLYEQSHIPHRMAYAACIGPGKHACQHSPHSTT
Summary
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Cofactor
Mg(2+)
Zn(2+)
Zn(2+)
Similarity
Belongs to the alkaline phosphatase family.
Keywords
Cell membrane
Complete proteome
Direct protein sequencing
Glycoprotein
GPI-anchor
Hydrolase
Lipoprotein
Magnesium
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Signal
Zinc
Feature
chain Alkaline phosphatase
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
B2ZZW8
B2ZZW4
Q7KXT0
C6L8N9
A5A747
B2ZZX2
+ More
H9JXN7 O96056 A0A0U2IR33 A0A2H1VP42 A0A2W1BMI5 A0A194Q2C7 A0A2A4J4Q0 A0A2H5CLJ8 I6PI83 A0A212F875 A0A194R1W7 I0E1Y2 A0A194R7D1 A0A194Q2F4 A0A2A4J355 A0A194R254 A0A2W1BHB3 C3U1V9 A0A2H5CLG8 A0A2H1WA01 G3GBT4 A0A0U1ZEU5 A0A109P4P6 B4Z1D7 C3U1W0 I6MML5 C3U1W1 A0A1B0RHM8 A0A0U1ZEC3 B4X983 B4Z1D6 A0A1B0RHM5 A0A0U2WPZ0 A0A2H1VQS9 A0A194Q8I8 C3U1W2 R4I4D0 G1JT78 A0A0U2WZB7 A0A0U1ZJ21 A0A194R2N2 M4M3L4 H9JH21 B2ZZX0 B2ZZW7 B2ZZX1 R4I4E9 P29523 A5A746 B2ZZW3 C6L8N8 H9JH27 B2ZZW5 A0A212F895 I6PF06 A0A2J7PHL4 A0A067QRJ2 A0A2W1BI04 A0A182JP07 A0A067QT98 A0A3G1LC34 A0A2L1IQA2 A0A1S4FLA4 A0A2H1WF77 A0A1Q3EV69 Q16WV8 A0A0P6JST2 A0A1Q3EV65 B0W3S9 C0JBW1 U5EGZ8 A0A182I2X2 Q7QJ58 A0A182WZ34 W8DT07 A0A182XYI9 A0A182KQV2 A0A182VAE3 A0A182SLH6 A0A182THI0 A0A182MT69 A0A182RTZ7 E0VEB3 A0A182JL64 A0A182QLW3 A0A182WJL0 A0A212FHB6 H9JK21 A0A0U1ZF85 A0A182NKM7 A0A182P207 A0A1J1ID93 A0A2H1V6P7 A0A1B0CIG9
H9JXN7 O96056 A0A0U2IR33 A0A2H1VP42 A0A2W1BMI5 A0A194Q2C7 A0A2A4J4Q0 A0A2H5CLJ8 I6PI83 A0A212F875 A0A194R1W7 I0E1Y2 A0A194R7D1 A0A194Q2F4 A0A2A4J355 A0A194R254 A0A2W1BHB3 C3U1V9 A0A2H5CLG8 A0A2H1WA01 G3GBT4 A0A0U1ZEU5 A0A109P4P6 B4Z1D7 C3U1W0 I6MML5 C3U1W1 A0A1B0RHM8 A0A0U1ZEC3 B4X983 B4Z1D6 A0A1B0RHM5 A0A0U2WPZ0 A0A2H1VQS9 A0A194Q8I8 C3U1W2 R4I4D0 G1JT78 A0A0U2WZB7 A0A0U1ZJ21 A0A194R2N2 M4M3L4 H9JH21 B2ZZX0 B2ZZW7 B2ZZX1 R4I4E9 P29523 A5A746 B2ZZW3 C6L8N8 H9JH27 B2ZZW5 A0A212F895 I6PF06 A0A2J7PHL4 A0A067QRJ2 A0A2W1BI04 A0A182JP07 A0A067QT98 A0A3G1LC34 A0A2L1IQA2 A0A1S4FLA4 A0A2H1WF77 A0A1Q3EV69 Q16WV8 A0A0P6JST2 A0A1Q3EV65 B0W3S9 C0JBW1 U5EGZ8 A0A182I2X2 Q7QJ58 A0A182WZ34 W8DT07 A0A182XYI9 A0A182KQV2 A0A182VAE3 A0A182SLH6 A0A182THI0 A0A182MT69 A0A182RTZ7 E0VEB3 A0A182JL64 A0A182QLW3 A0A182WJL0 A0A212FHB6 H9JK21 A0A0U1ZF85 A0A182NKM7 A0A182P207 A0A1J1ID93 A0A2H1V6P7 A0A1B0CIG9
EC Number
3.1.3.1
Pubmed
EMBL
AB379674
BAG41973.1
AB379672
BAG41969.1
AB055428
AB379673
+ More
BAB62746.2 BAG41971.1 AB516293 BAH95822.1 AB270698 BAF62125.1 AB379677 BAG41977.1 BABH01044837 AB013386 BAA34926.1 KT276304 ALS30431.1 ODYU01003612 SOQ42593.1 KZ150047 PZC74487.1 KQ459562 KPI99722.1 NWSH01003359 PCG66484.1 KY912922 AUH25163.1 JF979062 AFI81420.1 AGBW02009775 OWR49944.1 KQ460870 KPJ11778.1 JQ365182 AFH96950.1 KPJ11776.1 KPI99721.1 PCG66485.1 KPJ11777.1 PZC74489.1 FJ416470 ACP39712.1 KY912924 AUH25165.1 ODYU01007279 SOQ49911.1 JF825967 AEG79734.1 KM048196 AJW76723.1 KP420013 ALK86921.1 EU729323 ACF40807.1 FJ416471 ACP39713.1 HQ110096 AEL23233.1 FJ416472 ACP39714.1 KM360183 AKH49599.1 KM048195 AJW76722.1 EF531619 ABR88230.1 EU729322 ACF40806.1 KM360191 AKH49607.1 KT276297 ALS30424.1 SOQ42594.1 KPI99720.1 FJ416473 ACP39715.1 JN687589 AFJ04290.1 JN392445 AEM43806.1 KT276303 ALS30430.1 KM048197 AJW76724.1 KPJ11779.1 JQ747498 AGG36455.1 BABH01037520 AB379676 BAG41975.1 BAG41972.1 BAG41976.1 JN687588 AFJ04289.1 D90454 BAF62124.1 BAG41968.1 AB516292 BAH95821.1 BABH01037533 BABH01037534 BAG41970.1 OWR49943.1 JF979061 AFI81421.1 NEVH01025135 PNF15822.1 KK853042 KDR12134.1 PZC74488.1 KDR12135.1 KY922835 AUF74484.1 MF741672 AVD96955.1 ODYU01008153 SOQ51522.1 GFDL01015845 JAV19200.1 CH477553 EAT39089.1 GDUN01000056 JAN95863.1 GFDL01015846 JAV19199.1 DS231833 EDS32226.1 FJ175380 ACN29682.1 GANO01003229 JAB56642.1 APCN01000188 AAAB01008807 EAA03918.4 KC841472 AHF20243.2 AXCM01001173 DS235088 EEB11719.1 AXCN02000551 AGBW02008519 OWR53113.1 BABH01026539 BABH01026540 KM048198 AJW76725.1 CVRI01000047 CRK98187.1 ODYU01000769 SOQ36062.1 AJWK01013324
BAB62746.2 BAG41971.1 AB516293 BAH95822.1 AB270698 BAF62125.1 AB379677 BAG41977.1 BABH01044837 AB013386 BAA34926.1 KT276304 ALS30431.1 ODYU01003612 SOQ42593.1 KZ150047 PZC74487.1 KQ459562 KPI99722.1 NWSH01003359 PCG66484.1 KY912922 AUH25163.1 JF979062 AFI81420.1 AGBW02009775 OWR49944.1 KQ460870 KPJ11778.1 JQ365182 AFH96950.1 KPJ11776.1 KPI99721.1 PCG66485.1 KPJ11777.1 PZC74489.1 FJ416470 ACP39712.1 KY912924 AUH25165.1 ODYU01007279 SOQ49911.1 JF825967 AEG79734.1 KM048196 AJW76723.1 KP420013 ALK86921.1 EU729323 ACF40807.1 FJ416471 ACP39713.1 HQ110096 AEL23233.1 FJ416472 ACP39714.1 KM360183 AKH49599.1 KM048195 AJW76722.1 EF531619 ABR88230.1 EU729322 ACF40806.1 KM360191 AKH49607.1 KT276297 ALS30424.1 SOQ42594.1 KPI99720.1 FJ416473 ACP39715.1 JN687589 AFJ04290.1 JN392445 AEM43806.1 KT276303 ALS30430.1 KM048197 AJW76724.1 KPJ11779.1 JQ747498 AGG36455.1 BABH01037520 AB379676 BAG41975.1 BAG41972.1 BAG41976.1 JN687588 AFJ04289.1 D90454 BAF62124.1 BAG41968.1 AB516292 BAH95821.1 BABH01037533 BABH01037534 BAG41970.1 OWR49943.1 JF979061 AFI81421.1 NEVH01025135 PNF15822.1 KK853042 KDR12134.1 PZC74488.1 KDR12135.1 KY922835 AUF74484.1 MF741672 AVD96955.1 ODYU01008153 SOQ51522.1 GFDL01015845 JAV19200.1 CH477553 EAT39089.1 GDUN01000056 JAN95863.1 GFDL01015846 JAV19199.1 DS231833 EDS32226.1 FJ175380 ACN29682.1 GANO01003229 JAB56642.1 APCN01000188 AAAB01008807 EAA03918.4 KC841472 AHF20243.2 AXCM01001173 DS235088 EEB11719.1 AXCN02000551 AGBW02008519 OWR53113.1 BABH01026539 BABH01026540 KM048198 AJW76725.1 CVRI01000047 CRK98187.1 ODYU01000769 SOQ36062.1 AJWK01013324
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000235965
+ More
UP000027135 UP000075881 UP000008820 UP000002320 UP000075840 UP000007062 UP000076407 UP000076408 UP000075882 UP000075903 UP000075901 UP000075902 UP000075883 UP000075900 UP000009046 UP000075880 UP000075886 UP000075920 UP000075884 UP000075885 UP000183832 UP000092461
UP000027135 UP000075881 UP000008820 UP000002320 UP000075840 UP000007062 UP000076407 UP000076408 UP000075882 UP000075903 UP000075901 UP000075902 UP000075883 UP000075900 UP000009046 UP000075880 UP000075886 UP000075920 UP000075884 UP000075885 UP000183832 UP000092461
PRIDE
Pfam
PF00245 Alk_phosphatase
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
CDD
ProteinModelPortal
B2ZZW8
B2ZZW4
Q7KXT0
C6L8N9
A5A747
B2ZZX2
+ More
H9JXN7 O96056 A0A0U2IR33 A0A2H1VP42 A0A2W1BMI5 A0A194Q2C7 A0A2A4J4Q0 A0A2H5CLJ8 I6PI83 A0A212F875 A0A194R1W7 I0E1Y2 A0A194R7D1 A0A194Q2F4 A0A2A4J355 A0A194R254 A0A2W1BHB3 C3U1V9 A0A2H5CLG8 A0A2H1WA01 G3GBT4 A0A0U1ZEU5 A0A109P4P6 B4Z1D7 C3U1W0 I6MML5 C3U1W1 A0A1B0RHM8 A0A0U1ZEC3 B4X983 B4Z1D6 A0A1B0RHM5 A0A0U2WPZ0 A0A2H1VQS9 A0A194Q8I8 C3U1W2 R4I4D0 G1JT78 A0A0U2WZB7 A0A0U1ZJ21 A0A194R2N2 M4M3L4 H9JH21 B2ZZX0 B2ZZW7 B2ZZX1 R4I4E9 P29523 A5A746 B2ZZW3 C6L8N8 H9JH27 B2ZZW5 A0A212F895 I6PF06 A0A2J7PHL4 A0A067QRJ2 A0A2W1BI04 A0A182JP07 A0A067QT98 A0A3G1LC34 A0A2L1IQA2 A0A1S4FLA4 A0A2H1WF77 A0A1Q3EV69 Q16WV8 A0A0P6JST2 A0A1Q3EV65 B0W3S9 C0JBW1 U5EGZ8 A0A182I2X2 Q7QJ58 A0A182WZ34 W8DT07 A0A182XYI9 A0A182KQV2 A0A182VAE3 A0A182SLH6 A0A182THI0 A0A182MT69 A0A182RTZ7 E0VEB3 A0A182JL64 A0A182QLW3 A0A182WJL0 A0A212FHB6 H9JK21 A0A0U1ZF85 A0A182NKM7 A0A182P207 A0A1J1ID93 A0A2H1V6P7 A0A1B0CIG9
H9JXN7 O96056 A0A0U2IR33 A0A2H1VP42 A0A2W1BMI5 A0A194Q2C7 A0A2A4J4Q0 A0A2H5CLJ8 I6PI83 A0A212F875 A0A194R1W7 I0E1Y2 A0A194R7D1 A0A194Q2F4 A0A2A4J355 A0A194R254 A0A2W1BHB3 C3U1V9 A0A2H5CLG8 A0A2H1WA01 G3GBT4 A0A0U1ZEU5 A0A109P4P6 B4Z1D7 C3U1W0 I6MML5 C3U1W1 A0A1B0RHM8 A0A0U1ZEC3 B4X983 B4Z1D6 A0A1B0RHM5 A0A0U2WPZ0 A0A2H1VQS9 A0A194Q8I8 C3U1W2 R4I4D0 G1JT78 A0A0U2WZB7 A0A0U1ZJ21 A0A194R2N2 M4M3L4 H9JH21 B2ZZX0 B2ZZW7 B2ZZX1 R4I4E9 P29523 A5A746 B2ZZW3 C6L8N8 H9JH27 B2ZZW5 A0A212F895 I6PF06 A0A2J7PHL4 A0A067QRJ2 A0A2W1BI04 A0A182JP07 A0A067QT98 A0A3G1LC34 A0A2L1IQA2 A0A1S4FLA4 A0A2H1WF77 A0A1Q3EV69 Q16WV8 A0A0P6JST2 A0A1Q3EV65 B0W3S9 C0JBW1 U5EGZ8 A0A182I2X2 Q7QJ58 A0A182WZ34 W8DT07 A0A182XYI9 A0A182KQV2 A0A182VAE3 A0A182SLH6 A0A182THI0 A0A182MT69 A0A182RTZ7 E0VEB3 A0A182JL64 A0A182QLW3 A0A182WJL0 A0A212FHB6 H9JK21 A0A0U1ZF85 A0A182NKM7 A0A182P207 A0A1J1ID93 A0A2H1V6P7 A0A1B0CIG9
PDB
1SHQ
E-value=4.48472e-110,
Score=1019
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cell membrane
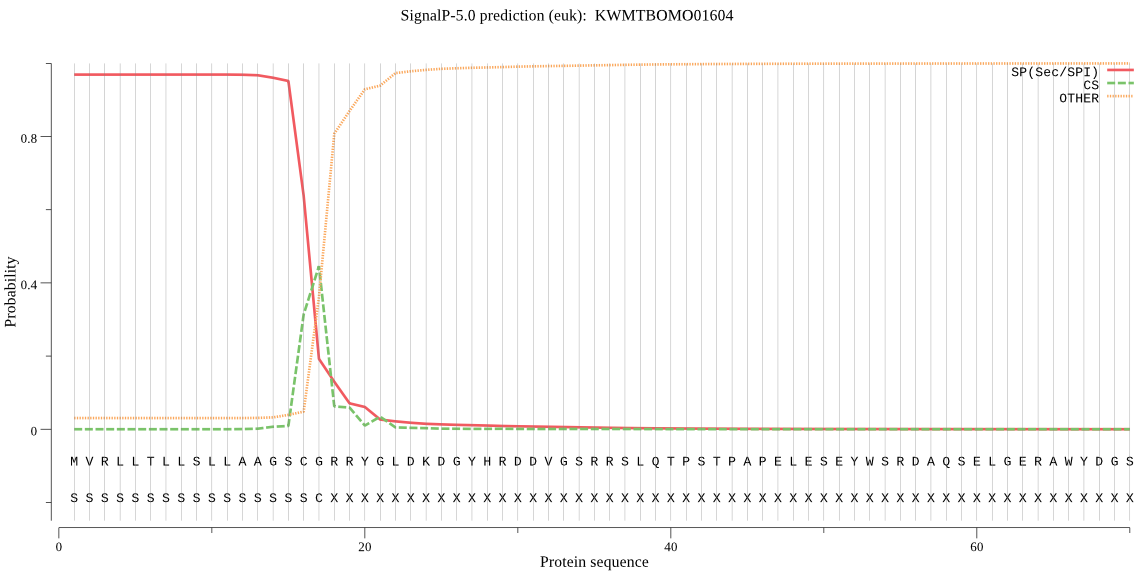
SignalP
Position: 1 - 17,
Likelihood: 0.969066
Length:
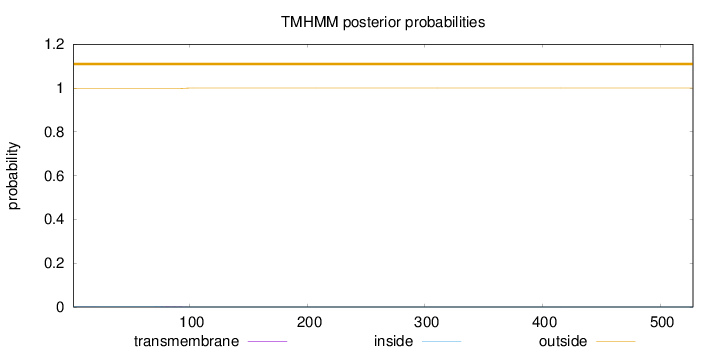
528
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0449400000000001
Exp number, first 60 AAs:
0.00696
Total prob of N-in:
0.00198
outside
1 - 528
Population Genetic Test Statistics
Pi
203.97141
Theta
176.83905
Tajima's D
0.403226
CLR
1.000838
CSRT
0.48267586620669
Interpretation
Uncertain
