Gene
KWMTBOMO01595
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.952 PlasmaMembrane Reliability : 1.288
Sequence
CDS
ATGCAATGTCCCAGCTGCATAAGATATGGAGAGACCAGAAAAACTAAAACCAAATTGGTGAGTTCGCTAGTTTTCTCCATATTCAATTATGGCGCGGAGACCTGGACAATGAAGAAGGCCGACAGAGACCGAATAGACGCTTTTGAAATGTGGTGTTGGAGAAAAATGCTACAGATTCCATGGACCGCCTTTCGCACCAACGTGTCTATTCTGAGAGAGCTTCATATCAAAGCCAGACTTTCCACTATATGCCTTCGTGGGGTGCTGGAATTCTTCGGACATATTGCTTGTAAAGAGGGTCACAATCTTGAACAGCTGATGGTGACAGGAAAGGTATATGGCAAACGTCCCAGAGAACGCAGTCCCACGAGATGGTCGGACCAAATACGATCTTCTCTGAGCATCAATTTCCATAATGCCCTTTATGAAGCAAAGGATCGCGGCAGATGGAGGGAAGTCGTATGGGAGAAACTGATGCAGCGGGGGAGTCTCGACCCTCAGTAA
Protein
MQCPSCIRYGETRKTKTKLVSSLVFSIFNYGAETWTMKKADRDRIDAFEMWCWRKMLQIPWTAFRTNVSILRELHIKARLSTICLRGVLEFFGHIACKEGHNLEQLMVTGKVYGKRPRERSPTRWSDQIRSSLSINFHNALYEAKDRGRWREVVWEKLMQRGSLDPQ
Summary
Uniprot
D7F163
A0A2H1VNK5
A0A2H1V742
A0A2H1VU13
D7F172
D7F161
+ More
D7F168 D7F162 D5LB39 D7F175 D7F167 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8JE23 H2YWZ6 A0A2G8K796 H2YWZ5 A0A2G8K327 A0A2G8KTJ4 A0A2G8JVA1 A0A2G8L6P4 W5NMM5 A0A2G8K183 A0A2G8JVI4 O97916 W5NIE2 X1D4L4 W4Y921 W5MM77 A0A2H6KKM9 A0A085LZ21 W4YPU3 A0A2A4J795 A0A061BKJ9 W5MW18 W5NLK6 J9LZK0 X1WK45 A0A0B7BSZ9 A0A0B7BUR9 A0A0B7BT80 W5N9W4 A0A067RQM5 A0A067QJX4 A0A067QUH9 A0A067QZY5 A0A0B7BVD9 W5NMX6 A0A0B7BVD3 A0A0B7BSB6 A0A067R8J7 W5MYT3 A0A067QXS5 A0A146L0Z1 A0A2W1BSS7 W4YNG7 W4ZJA4 A0A067RDT2 A0A067RS42 D5LB38 A0A0B7BT11 A0A2S2NMQ2 X1XC23 A0A0J7KZE8 W4XFH1 A0A076N3Z4 A0A0K8T3U8 H3B2Y3
D7F168 D7F162 D5LB39 D7F175 D7F167 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8JE23 H2YWZ6 A0A2G8K796 H2YWZ5 A0A2G8K327 A0A2G8KTJ4 A0A2G8JVA1 A0A2G8L6P4 W5NMM5 A0A2G8K183 A0A2G8JVI4 O97916 W5NIE2 X1D4L4 W4Y921 W5MM77 A0A2H6KKM9 A0A085LZ21 W4YPU3 A0A2A4J795 A0A061BKJ9 W5MW18 W5NLK6 J9LZK0 X1WK45 A0A0B7BSZ9 A0A0B7BUR9 A0A0B7BT80 W5N9W4 A0A067RQM5 A0A067QJX4 A0A067QUH9 A0A067QZY5 A0A0B7BVD9 W5NMX6 A0A0B7BVD3 A0A0B7BSB6 A0A067R8J7 W5MYT3 A0A067QXS5 A0A146L0Z1 A0A2W1BSS7 W4YNG7 W4ZJA4 A0A067RDT2 A0A067RS42 D5LB38 A0A0B7BT11 A0A2S2NMQ2 X1XC23 A0A0J7KZE8 W4XFH1 A0A076N3Z4 A0A0K8T3U8 H3B2Y3
Pubmed
EMBL
FJ265548
ADI61816.1
ODYU01003523
SOQ42419.1
ODYU01001007
SOQ36619.1
+ More
ODYU01004401 SOQ44236.1 FJ265557 ADI61825.1 FJ265546 ADI61814.1 FJ265553 ADI61821.1 FJ265547 ADI61815.1 GU815090 ADF18553.1 FJ265560 ADI61828.1 FJ265552 ADI61820.1 KZ150203 PZC72267.1 FJ265559 ADI61827.1 MRZV01000056 PIK60437.1 MRZV01002322 PIK33998.1 MRZV01000817 PIK43880.1 MRZV01000940 PIK42365.1 MRZV01000378 PIK51329.1 MRZV01001216 PIK39650.1 MRZV01000196 PIK55924.1 AHAT01022938 MRZV01000994 PIK41715.1 MRZV01001207 PIK39730.1 AJ132772 CAA10770.1 AHAT01009145 BART01031562 GAH15691.1 AAGJ04161766 AHAT01015286 BDSA01000081 GBE63537.1 KL363256 KL367985 KFD50217.1 KFD59415.1 AAGJ04055194 NWSH01002736 PCG67606.1 LK055282 CDR71995.1 AHAT01013157 AHAT01013158 AHAT01012943 ABLF02042206 ABLF02024992 ABLF02024994 ABLF02025002 ABLF02043975 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 AHAT01022314 KK852482 KDR22935.1 KK853249 KDR09341.1 KK852941 KDR13536.1 KK852843 KDR15156.1 HACG01049255 CEK96120.1 AHAT01025955 HACG01049245 CEK96110.1 HACG01049249 CEK96114.1 KK852811 KDR15914.1 AHAT01017643 KK853131 KDR10935.1 GDHC01017214 JAQ01415.1 KZ149985 PZC75696.1 AAGJ04167631 AAGJ04021158 KK852530 KDR21932.1 KK852456 KDR23625.1 GU815089 ADF18552.1 HACG01049494 HACG01049495 CEK96359.1 CEK96360.1 GGMR01005851 MBY18470.1 ABLF02031909 LBMM01001656 KMQ95932.1 AAGJ04075245 KF881086 AIJ27485.1 GBRD01005612 JAG60209.1 AFYH01119920
ODYU01004401 SOQ44236.1 FJ265557 ADI61825.1 FJ265546 ADI61814.1 FJ265553 ADI61821.1 FJ265547 ADI61815.1 GU815090 ADF18553.1 FJ265560 ADI61828.1 FJ265552 ADI61820.1 KZ150203 PZC72267.1 FJ265559 ADI61827.1 MRZV01000056 PIK60437.1 MRZV01002322 PIK33998.1 MRZV01000817 PIK43880.1 MRZV01000940 PIK42365.1 MRZV01000378 PIK51329.1 MRZV01001216 PIK39650.1 MRZV01000196 PIK55924.1 AHAT01022938 MRZV01000994 PIK41715.1 MRZV01001207 PIK39730.1 AJ132772 CAA10770.1 AHAT01009145 BART01031562 GAH15691.1 AAGJ04161766 AHAT01015286 BDSA01000081 GBE63537.1 KL363256 KL367985 KFD50217.1 KFD59415.1 AAGJ04055194 NWSH01002736 PCG67606.1 LK055282 CDR71995.1 AHAT01013157 AHAT01013158 AHAT01012943 ABLF02042206 ABLF02024992 ABLF02024994 ABLF02025002 ABLF02043975 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 AHAT01022314 KK852482 KDR22935.1 KK853249 KDR09341.1 KK852941 KDR13536.1 KK852843 KDR15156.1 HACG01049255 CEK96120.1 AHAT01025955 HACG01049245 CEK96110.1 HACG01049249 CEK96114.1 KK852811 KDR15914.1 AHAT01017643 KK853131 KDR10935.1 GDHC01017214 JAQ01415.1 KZ149985 PZC75696.1 AAGJ04167631 AAGJ04021158 KK852530 KDR21932.1 KK852456 KDR23625.1 GU815089 ADF18552.1 HACG01049494 HACG01049495 CEK96359.1 CEK96360.1 GGMR01005851 MBY18470.1 ABLF02031909 LBMM01001656 KMQ95932.1 AAGJ04075245 KF881086 AIJ27485.1 GBRD01005612 JAG60209.1 AFYH01119920
Proteomes
Interpro
Gene 3D
ProteinModelPortal
D7F163
A0A2H1VNK5
A0A2H1V742
A0A2H1VU13
D7F172
D7F161
+ More
D7F168 D7F162 D5LB39 D7F175 D7F167 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8JE23 H2YWZ6 A0A2G8K796 H2YWZ5 A0A2G8K327 A0A2G8KTJ4 A0A2G8JVA1 A0A2G8L6P4 W5NMM5 A0A2G8K183 A0A2G8JVI4 O97916 W5NIE2 X1D4L4 W4Y921 W5MM77 A0A2H6KKM9 A0A085LZ21 W4YPU3 A0A2A4J795 A0A061BKJ9 W5MW18 W5NLK6 J9LZK0 X1WK45 A0A0B7BSZ9 A0A0B7BUR9 A0A0B7BT80 W5N9W4 A0A067RQM5 A0A067QJX4 A0A067QUH9 A0A067QZY5 A0A0B7BVD9 W5NMX6 A0A0B7BVD3 A0A0B7BSB6 A0A067R8J7 W5MYT3 A0A067QXS5 A0A146L0Z1 A0A2W1BSS7 W4YNG7 W4ZJA4 A0A067RDT2 A0A067RS42 D5LB38 A0A0B7BT11 A0A2S2NMQ2 X1XC23 A0A0J7KZE8 W4XFH1 A0A076N3Z4 A0A0K8T3U8 H3B2Y3
D7F168 D7F162 D5LB39 D7F175 D7F167 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8JE23 H2YWZ6 A0A2G8K796 H2YWZ5 A0A2G8K327 A0A2G8KTJ4 A0A2G8JVA1 A0A2G8L6P4 W5NMM5 A0A2G8K183 A0A2G8JVI4 O97916 W5NIE2 X1D4L4 W4Y921 W5MM77 A0A2H6KKM9 A0A085LZ21 W4YPU3 A0A2A4J795 A0A061BKJ9 W5MW18 W5NLK6 J9LZK0 X1WK45 A0A0B7BSZ9 A0A0B7BUR9 A0A0B7BT80 W5N9W4 A0A067RQM5 A0A067QJX4 A0A067QUH9 A0A067QZY5 A0A0B7BVD9 W5NMX6 A0A0B7BVD3 A0A0B7BSB6 A0A067R8J7 W5MYT3 A0A067QXS5 A0A146L0Z1 A0A2W1BSS7 W4YNG7 W4ZJA4 A0A067RDT2 A0A067RS42 D5LB38 A0A0B7BT11 A0A2S2NMQ2 X1XC23 A0A0J7KZE8 W4XFH1 A0A076N3Z4 A0A0K8T3U8 H3B2Y3
Ontologies
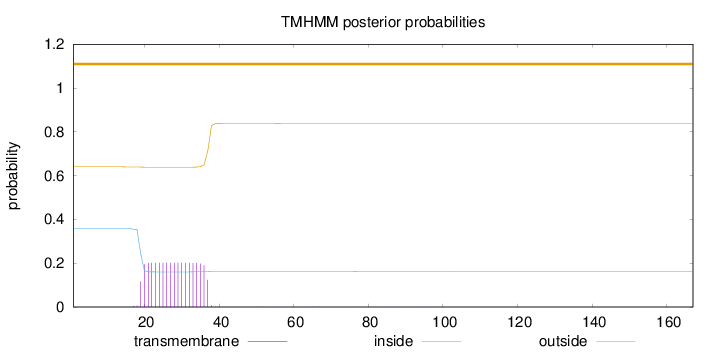
Topology
Length:
167
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.68711
Exp number, first 60 AAs:
3.66687
Total prob of N-in:
0.35985
outside
1 - 167
Population Genetic Test Statistics
Pi
160.028609
Theta
129.471277
Tajima's D
0.741312
CLR
0
CSRT
0.583470826458677
Interpretation
Uncertain
