Gene
KWMTBOMO01593
Pre Gene Modal
BGIBMGA008827
Annotation
serine_protease_inhibitor_20_precursor_[Bombyx_mori]
Full name
Alaserpin
+ More
Serine protease inhibitor 3/4
Antichymotrypsin-2
Antitrypsin
Serine protease inhibitor 3/4
Antichymotrypsin-2
Antitrypsin
Alternative Name
Serpin-1
Antichymotrypsin II
Antichymotrypsin II
Location in the cell
Nuclear Reliability : 3.858
Sequence
CDS
ATGGAATATTACTACAGTTTCGTCACGATTGTGTTCCCGCTGCTAAGCGCATTGGGTCAGCTGGCCTTGTTCTCGGAAAAGAAAAACGAACAAAAGCTTCTTGATCTTCTAGAATTGGATAATAAGGAACAGATACGTAGAATATTTCCGTATATACGAGAGGAGTACTTCAGTGAAGACCTTGTAGCTTCACCGAAATTCGACCTGAAAGTTTACACTGACGACGACAACAAGCTGTGTCCCGCTTTCAAAAAGTCATACAAAAAGACCTTTGGTGGTGAGACCGGGGAGGTAGACTACAGTGAACCCGAAGAAGCTGCTGAAGAAATCAACGATTATGTAAAAAGTCAAGGCACAGGTGGCTTCAAAGACCTCGTTTCAAGTCGAAAAATAGAGGACAAGGATGGACTGAATTTTATAGGCACCTACGAGCTAAGTTTTAAATTTGACGATAGATTCGTTTTCACGAGCAGCAAAGTGGTGAAGTTCAAAACCGGTAAGACTGTTACAATCATTCCCGGGGCGGCCGGGGAAGGTATCATCAAATATGCCGACGTTAAATCTATAAACGCCAAGGTTATAGAAATTCTGGCTAGAGGTGGAGAATACTCGTATGTCGTAGTTCTTCCAAACAAAGTTGATGGACTGAAAGAGGTACTGAAGAAATTCACAGATCCTGAAGTTTTCAGACAAGCCATAGACCAAGGGAAAAACGTTTGTGCCAAACTATTAGTTCCCAGCCGAGAAGTGATCAGTAAGGTGGATTTAGCAAACTCTCTCCAACAGTTCCGTTTTGATGAAGCCGATTCCTTTATCGTGATAACATTGAGAGAAGAAATATTCAACAGATGGGAACCAAATGAAACCGGAACCTTCTTGCAGTGA
Protein
MEYYYSFVTIVFPLLSALGQLALFSEKKNEQKLLDLLELDNKEQIRRIFPYIREEYFSEDLVASPKFDLKVYTDDDNKLCPAFKKSYKKTFGGETGEVDYSEPEEAAEEINDYVKSQGTGGFKDLVSSRKIEDKDGLNFIGTYELSFKFDDRFVFTSSKVVKFKTGKTVTIIPGAAGEGIIKYADVKSINAKVIEILARGGEYSYVVVLPNKVDGLKEVLKKFTDPEVFRQAIDQGKNVCAKLLVPSREVISKVDLANSLQQFRFDEADSFIVITLREEIFNRWEPNETGTFLQ
Summary
Miscellaneous
The N-terminal section (1-336) is identical to the N-terminal section of the silk moth antitrypsin I (17-352).
Similarity
Belongs to the serpin family.
Keywords
3D-structure
Alternative splicing
Glycoprotein
Protease inhibitor
Secreted
Serine protease inhibitor
Signal
Direct protein sequencing
Complete proteome
Reference proteome
Feature
chain Alaserpin
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JH30
C0J8G9
C0J8H3
C0J8G4
C0J8G6
H9JH34
+ More
H9JH33 C0J8H9 C0J8H4 C0J8H2 C0J8G8 H9J5E5 A0A3G1T1Q1 H9J5E6 A0A3G1T1R3 F5B4G8 G9F9J2 B4XHA0 H9JH29 B4XHA1 B4XH99 B4XH98 A0A385XRR7 G9F9J3 A0A385XRT7 Q25499 Q25494 Q25501 Q25495 A0A2H1VE36 Q25497 P14754 Q25526 Q25492 Q25491 Q25493 Q25498 Q25496 Q25500 Q5MGH0 D9HQ60 A0A1L7B975 K4P392 D9HQA1 G9D4T8 A0A0L7LMC8 B0FZ64 B0FZ60 B0FZ62 B0FZ63 B0FZ66 B0FZ68 B0FZ65 B0FZ67 B0FZ61 Q8IS84 Q8IS85 Q8IS86 C5I794 C5I795 A3KEZ8 Q60FS0 A3KEZ7 B2X122 P80034 C7ASM2 P22922 C7ASM3 C7ASM4 A0A0M4K7J2 A0A212FD15
H9JH33 C0J8H9 C0J8H4 C0J8H2 C0J8G8 H9J5E5 A0A3G1T1Q1 H9J5E6 A0A3G1T1R3 F5B4G8 G9F9J2 B4XHA0 H9JH29 B4XHA1 B4XH99 B4XH98 A0A385XRR7 G9F9J3 A0A385XRT7 Q25499 Q25494 Q25501 Q25495 A0A2H1VE36 Q25497 P14754 Q25526 Q25492 Q25491 Q25493 Q25498 Q25496 Q25500 Q5MGH0 D9HQ60 A0A1L7B975 K4P392 D9HQA1 G9D4T8 A0A0L7LMC8 B0FZ64 B0FZ60 B0FZ62 B0FZ63 B0FZ66 B0FZ68 B0FZ65 B0FZ67 B0FZ61 Q8IS84 Q8IS85 Q8IS86 C5I794 C5I795 A3KEZ8 Q60FS0 A3KEZ7 B2X122 P80034 C7ASM2 P22922 C7ASM3 C7ASM4 A0A0M4K7J2 A0A212FD15
Pubmed
EMBL
BABH01037565
EU935621
ACG61183.1
EU935625
ACG61187.1
EU935616
+ More
ACG61178.1 EU935618 ACG61180.1 BABH01037571 BABH01037572 BABH01037570 EU935631 ACG61193.1 EU935626 ACG61188.1 EU935624 ACG61186.1 EU935620 ACG61182.1 BABH01039199 MG770320 AXY94922.1 BABH01039200 MG770321 AXY94923.1 HQ615869 ADT63775.1 JN033737 AEW46890.2 EU040209 ABW17157.1 BABH01037558 EU040210 ABW17158.1 EU040208 ABW17156.1 EU040207 ABW17155.1 MG753799 AYC12556.1 JN033738 AEW46891.2 MG753798 AYC12555.1 U58361 AAC47340.1 AAC47332.1 AAC47339.1 AAC47341.1 ODYU01002062 SOQ39103.1 AAC47333.1 M23438 L20792 L20793 AAA29334.1 AAC47338.1 AAC47334.1 AAC47337.1 AAC47342.1 AAC47336.1 AAC47335.1 AY829816 AY829817 AAV91430.1 HM023815 JF777348 ADJ58548.1 AEU11693.1 KY385635 APT69916.1 JX524148 AFV46312.1 HM023856 ADJ58589.2 JF777341 AEU11686.1 JTDY01000556 KOB76688.1 EU368832 ABY68559.1 ABY68563.1 ABY68561.1 ABY68560.1 ABY68557.1 ABY68555.1 ABY68558.1 ABY68556.1 ABY68562.1 AY148485 AAN71634.1 AY148484 AAN71633.1 AY148483 AAN71632.1 FJ763763 ACR56864.1 FJ763764 ACR56865.1 AB295499 BAF48335.1 AB189029 BAD52261.1 AB295498 BAF48334.1 EF584499 ABU62829.1 FJ613793 ACT36272.1 D00738 FJ613794 FJ613796 ACT36273.1 ACT36275.1 FJ613795 ACT36274.1 KR003729 ALD62507.1 AGBW02009121 OWR51607.1
ACG61178.1 EU935618 ACG61180.1 BABH01037571 BABH01037572 BABH01037570 EU935631 ACG61193.1 EU935626 ACG61188.1 EU935624 ACG61186.1 EU935620 ACG61182.1 BABH01039199 MG770320 AXY94922.1 BABH01039200 MG770321 AXY94923.1 HQ615869 ADT63775.1 JN033737 AEW46890.2 EU040209 ABW17157.1 BABH01037558 EU040210 ABW17158.1 EU040208 ABW17156.1 EU040207 ABW17155.1 MG753799 AYC12556.1 JN033738 AEW46891.2 MG753798 AYC12555.1 U58361 AAC47340.1 AAC47332.1 AAC47339.1 AAC47341.1 ODYU01002062 SOQ39103.1 AAC47333.1 M23438 L20792 L20793 AAA29334.1 AAC47338.1 AAC47334.1 AAC47337.1 AAC47342.1 AAC47336.1 AAC47335.1 AY829816 AY829817 AAV91430.1 HM023815 JF777348 ADJ58548.1 AEU11693.1 KY385635 APT69916.1 JX524148 AFV46312.1 HM023856 ADJ58589.2 JF777341 AEU11686.1 JTDY01000556 KOB76688.1 EU368832 ABY68559.1 ABY68563.1 ABY68561.1 ABY68560.1 ABY68557.1 ABY68555.1 ABY68558.1 ABY68556.1 ABY68562.1 AY148485 AAN71634.1 AY148484 AAN71633.1 AY148483 AAN71632.1 FJ763763 ACR56864.1 FJ763764 ACR56865.1 AB295499 BAF48335.1 AB189029 BAD52261.1 AB295498 BAF48334.1 EF584499 ABU62829.1 FJ613793 ACT36272.1 D00738 FJ613794 FJ613796 ACT36273.1 ACT36275.1 FJ613795 ACT36274.1 KR003729 ALD62507.1 AGBW02009121 OWR51607.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JH30
C0J8G9
C0J8H3
C0J8G4
C0J8G6
H9JH34
+ More
H9JH33 C0J8H9 C0J8H4 C0J8H2 C0J8G8 H9J5E5 A0A3G1T1Q1 H9J5E6 A0A3G1T1R3 F5B4G8 G9F9J2 B4XHA0 H9JH29 B4XHA1 B4XH99 B4XH98 A0A385XRR7 G9F9J3 A0A385XRT7 Q25499 Q25494 Q25501 Q25495 A0A2H1VE36 Q25497 P14754 Q25526 Q25492 Q25491 Q25493 Q25498 Q25496 Q25500 Q5MGH0 D9HQ60 A0A1L7B975 K4P392 D9HQA1 G9D4T8 A0A0L7LMC8 B0FZ64 B0FZ60 B0FZ62 B0FZ63 B0FZ66 B0FZ68 B0FZ65 B0FZ67 B0FZ61 Q8IS84 Q8IS85 Q8IS86 C5I794 C5I795 A3KEZ8 Q60FS0 A3KEZ7 B2X122 P80034 C7ASM2 P22922 C7ASM3 C7ASM4 A0A0M4K7J2 A0A212FD15
H9JH33 C0J8H9 C0J8H4 C0J8H2 C0J8G8 H9J5E5 A0A3G1T1Q1 H9J5E6 A0A3G1T1R3 F5B4G8 G9F9J2 B4XHA0 H9JH29 B4XHA1 B4XH99 B4XH98 A0A385XRR7 G9F9J3 A0A385XRT7 Q25499 Q25494 Q25501 Q25495 A0A2H1VE36 Q25497 P14754 Q25526 Q25492 Q25491 Q25493 Q25498 Q25496 Q25500 Q5MGH0 D9HQ60 A0A1L7B975 K4P392 D9HQA1 G9D4T8 A0A0L7LMC8 B0FZ64 B0FZ60 B0FZ62 B0FZ63 B0FZ66 B0FZ68 B0FZ65 B0FZ67 B0FZ61 Q8IS84 Q8IS85 Q8IS86 C5I794 C5I795 A3KEZ8 Q60FS0 A3KEZ7 B2X122 P80034 C7ASM2 P22922 C7ASM3 C7ASM4 A0A0M4K7J2 A0A212FD15
PDB
1K9O
E-value=3.15599e-14,
Score=189
Ontologies
PANTHER
Topology
Subcellular location
Secreted
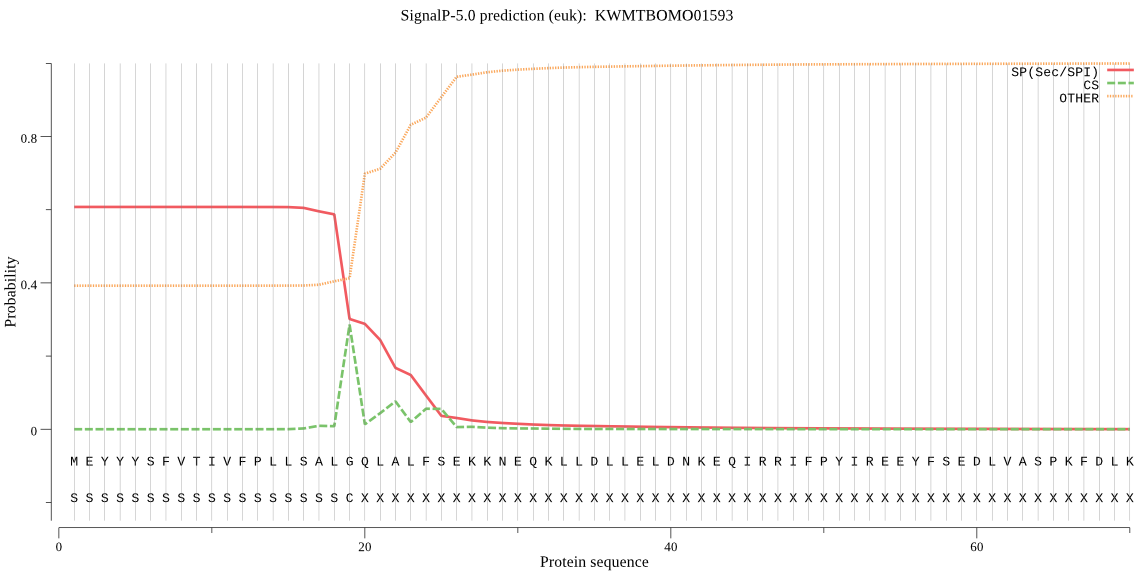
SignalP
Position: 1 - 19,
Likelihood: 0.606647
Length:
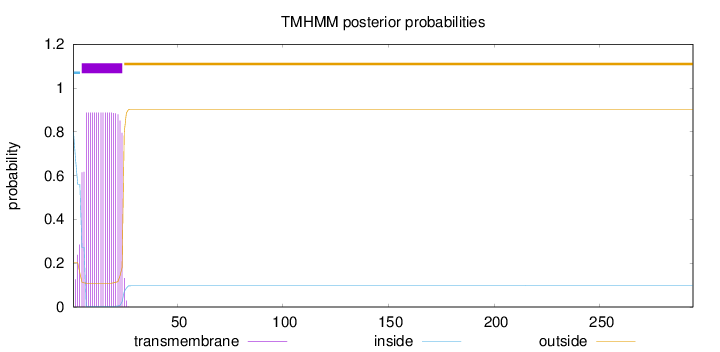
294
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.88688
Exp number, first 60 AAs:
17.88473
Total prob of N-in:
0.79846
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 24
outside
25 - 294
Population Genetic Test Statistics
Pi
315.008297
Theta
207.023008
Tajima's D
1.827767
CLR
20.686834
CSRT
0.855957202139893
Interpretation
Uncertain
