Gene
KWMTBOMO01592 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008829
Annotation
PREDICTED:_alaserpin-like_[Bombyx_mori]
Full name
Alaserpin
Alternative Name
Serpin-1
Location in the cell
Mitochondrial Reliability : 2.401
Sequence
CDS
ATGTACAGTGATACGAGCAACAGGTTAAGTCCAGCTTTCATAGATACCTACAACAAAACCTTTGGTGGTAAGGCTAGGACTGTGGACTACAGTGACACCGTTAAAGCTGCCAATGAGATCAATGATGAGGTTGAAAATGAAGGAGAGTTCAATAATATAATCTCCAGTAAAATGATAAAGGAAGGAGGCGGTCTTAGTTTTATAAGCAGCTACGTGTTGACTATCATGTACAGCGACAAATTCAAATTCATAAACGAGAAAACAATAAACTTCAAATCTGGTAACAAGGTGATAAATATTCCAGGAATTACCGGGGAAGGTGTAGTGAAATATGCCGACATAAAGTCTATTGATGCCAAGACCATAGAATTGGAAGGTAAAGATGGACTATACTCGTATATCATGGTCCTTCCAAACAAACATGATGGACTTCTGGAAGTACTAGAAAAGCTCTGTGATTCTACAGCTTTCCAAGAAATTCTTGATCGTGGCGAGGAAGTATGCGCAAAATTATCCATCCCAGCCGGAGACTTGATTGCCAAATGTGACTTAGAAAAGACAGCTAAAGAATCAAATTTATTAAGAGATATATTCGAAGAGGGGAGACTCGAACTCCAAGGTGTGAGTCATAAATATAATAATACATACGTTTCGTCAATCCTACATGAAGTTCATTTGAAGCCAGGTCCGATCATTTACGATGAGCCCAGAGAATTATGTTCTATGCCTCAAATAGAACTGGAGATGAACCGTCCTCATTTGTTCTACGTGACTTTCACACAGAAGGTTGGCCCGATCCGTACCCCAATAGTCGGTGTAGTGTTTGACATCACGGGTACAACAAGCAACTAG
Protein
MYSDTSNRLSPAFIDTYNKTFGGKARTVDYSDTVKAANEINDEVENEGEFNNIISSKMIKEGGGLSFISSYVLTIMYSDKFKFINEKTINFKSGNKVINIPGITGEGVVKYADIKSIDAKTIELEGKDGLYSYIMVLPNKHDGLLEVLEKLCDSTAFQEILDRGEEVCAKLSIPAGDLIAKCDLEKTAKESNLLRDIFEEGRLELQGVSHKYNNTYVSSILHEVHLKPGPIIYDEPRELCSMPQIELEMNRPHLFYVTFTQKVGPIRTPIVGVVFDITGTTSN
Summary
Similarity
Belongs to the serpin family.
Keywords
3D-structure
Alternative splicing
Glycoprotein
Protease inhibitor
Secreted
Serine protease inhibitor
Signal
Feature
chain Alaserpin
Uniprot
C0J8H3
H9JH32
C0J8G4
H9JH34
C0J8G6
H9JH33
+ More
C0J8G9 H9JH30 C0J8H4 C0J8H2 H9J5E5 C0J8H9 C0J8G8 Q25495 Q25494 A0A3G1T1R3 Q25496 G9F9J2 A0A3G1T1Q1 Q7JQ70 Q25500 Q25526 P14754 Q25501 Q25491 Q25492 Q25499 Q25497 Q25498 Q25493 A0A385XRT7 A0A385XRR7 A0A1B6KER4 A0A1B6K9M0 G9F9J3 A0A1B6LVE8 A0A212EIQ3 A0A1B6MQA5 W5ZZB8 A0A1B6L1S1 A0A1B6L303 A0A1B6IT90 W8C425 A0A0A0U6U7 A0A2W1BH42 F5B4G8 H9J5E6 A0A1B6IUV9 V5WCC7 A0A1B6HES2 A0A1B6J4R9 B0FZ67 A0A0P4WD07 A0A0P4W1G3 A0A1B6I5M5 A0A0N9EI66 A0A0P4W9M9 A0A0P4W7J0 A0A0N7ZBP8 A0A0A1X6Z0 A0A0A1WVW1 B4XHA1 A0A0A1XI57 A0A2P8YBQ0 B4XHA0 A0A0R1DLU4 A0A0R1DT85 A0A0R1DR25 A0A0P4WLT4 A0A0R1DSU2 A0A0R1DSX8 B4XH99 A0A1B6FM52 A0A059VDA6 B0FZ64 B0FZ62 B0FZ66 B0FZ63 B0FZ65 B0FZ68 A0A0L7LMC8
C0J8G9 H9JH30 C0J8H4 C0J8H2 H9J5E5 C0J8H9 C0J8G8 Q25495 Q25494 A0A3G1T1R3 Q25496 G9F9J2 A0A3G1T1Q1 Q7JQ70 Q25500 Q25526 P14754 Q25501 Q25491 Q25492 Q25499 Q25497 Q25498 Q25493 A0A385XRT7 A0A385XRR7 A0A1B6KER4 A0A1B6K9M0 G9F9J3 A0A1B6LVE8 A0A212EIQ3 A0A1B6MQA5 W5ZZB8 A0A1B6L1S1 A0A1B6L303 A0A1B6IT90 W8C425 A0A0A0U6U7 A0A2W1BH42 F5B4G8 H9J5E6 A0A1B6IUV9 V5WCC7 A0A1B6HES2 A0A1B6J4R9 B0FZ67 A0A0P4WD07 A0A0P4W1G3 A0A1B6I5M5 A0A0N9EI66 A0A0P4W9M9 A0A0P4W7J0 A0A0N7ZBP8 A0A0A1X6Z0 A0A0A1WVW1 B4XHA1 A0A0A1XI57 A0A2P8YBQ0 B4XHA0 A0A0R1DLU4 A0A0R1DT85 A0A0R1DR25 A0A0P4WLT4 A0A0R1DSU2 A0A0R1DSX8 B4XH99 A0A1B6FM52 A0A059VDA6 B0FZ64 B0FZ62 B0FZ66 B0FZ63 B0FZ65 B0FZ68 A0A0L7LMC8
Pubmed
EMBL
EU935625
ACG61187.1
BABH01037567
BABH01037568
BABH01037569
BABH01037570
+ More
EU935616 ACG61178.1 BABH01037571 BABH01037572 EU935618 ACG61180.1 EU935621 ACG61183.1 BABH01037565 EU935626 ACG61188.1 EU935624 ACG61186.1 BABH01039199 EU935631 ACG61193.1 EU935620 ACG61182.1 U58361 AAC47341.1 AAC47332.1 MG770321 AXY94923.1 AAC47336.1 JN033737 AEW46890.2 MG770320 AXY94922.1 L20790 AAA29328.1 AAC47335.1 L20793 AAA29334.1 M23438 L20792 AAC47339.1 AAC47334.1 AAC47338.1 AAC47340.1 AAC47333.1 AAC47342.1 AAC47337.1 MG753798 AYC12555.1 MG753799 AYC12556.1 GEBQ01030062 JAT09915.1 GEBQ01031830 JAT08147.1 JN033738 AEW46891.2 GEBQ01012324 JAT27653.1 AGBW02014593 OWR41373.1 GEBQ01001894 JAT38083.1 KF873763 AHI48498.1 GEBQ01022377 JAT17600.1 GEBQ01021952 JAT18025.1 GECU01017579 JAS90127.1 GAMC01005784 JAC00772.1 KM280385 AIW39913.1 KZ150134 PZC73004.1 HQ615869 ADT63775.1 BABH01039200 GECU01016985 JAS90721.1 KC577446 AHC06147.1 GECU01034479 JAS73227.1 GECU01013546 JAS94160.1 EU368832 ABY68556.1 GDRN01080305 JAI62250.1 GDRN01080302 JAI62252.1 GECU01025480 JAS82226.1 KR140160 ALF44673.1 GDRN01080301 JAI62253.1 GDRN01080303 JAI62251.1 GDRN01080307 JAI62248.1 GBXI01007869 JAD06423.1 GBXI01011724 JAD02568.1 EU040210 ABW17158.1 GBXI01004114 JAD10178.1 PYGN01000723 PSN41681.1 EU040209 ABW17157.1 CM000157 KRJ98200.1 KRJ98201.1 KRJ98195.1 KRJ98194.1 GDRN01080306 JAI62249.1 KRJ98192.1 KRJ98193.1 EU040208 ABW17156.1 GECZ01018515 GECZ01006882 JAS51254.1 JAS62887.1 KJ489357 AHZ96593.1 ABY68559.1 ABY68561.1 ABY68557.1 ABY68560.1 ABY68558.1 ABY68555.1 JTDY01000556 KOB76688.1
EU935616 ACG61178.1 BABH01037571 BABH01037572 EU935618 ACG61180.1 EU935621 ACG61183.1 BABH01037565 EU935626 ACG61188.1 EU935624 ACG61186.1 BABH01039199 EU935631 ACG61193.1 EU935620 ACG61182.1 U58361 AAC47341.1 AAC47332.1 MG770321 AXY94923.1 AAC47336.1 JN033737 AEW46890.2 MG770320 AXY94922.1 L20790 AAA29328.1 AAC47335.1 L20793 AAA29334.1 M23438 L20792 AAC47339.1 AAC47334.1 AAC47338.1 AAC47340.1 AAC47333.1 AAC47342.1 AAC47337.1 MG753798 AYC12555.1 MG753799 AYC12556.1 GEBQ01030062 JAT09915.1 GEBQ01031830 JAT08147.1 JN033738 AEW46891.2 GEBQ01012324 JAT27653.1 AGBW02014593 OWR41373.1 GEBQ01001894 JAT38083.1 KF873763 AHI48498.1 GEBQ01022377 JAT17600.1 GEBQ01021952 JAT18025.1 GECU01017579 JAS90127.1 GAMC01005784 JAC00772.1 KM280385 AIW39913.1 KZ150134 PZC73004.1 HQ615869 ADT63775.1 BABH01039200 GECU01016985 JAS90721.1 KC577446 AHC06147.1 GECU01034479 JAS73227.1 GECU01013546 JAS94160.1 EU368832 ABY68556.1 GDRN01080305 JAI62250.1 GDRN01080302 JAI62252.1 GECU01025480 JAS82226.1 KR140160 ALF44673.1 GDRN01080301 JAI62253.1 GDRN01080303 JAI62251.1 GDRN01080307 JAI62248.1 GBXI01007869 JAD06423.1 GBXI01011724 JAD02568.1 EU040210 ABW17158.1 GBXI01004114 JAD10178.1 PYGN01000723 PSN41681.1 EU040209 ABW17157.1 CM000157 KRJ98200.1 KRJ98201.1 KRJ98195.1 KRJ98194.1 GDRN01080306 JAI62249.1 KRJ98192.1 KRJ98193.1 EU040208 ABW17156.1 GECZ01018515 GECZ01006882 JAS51254.1 JAS62887.1 KJ489357 AHZ96593.1 ABY68559.1 ABY68561.1 ABY68557.1 ABY68560.1 ABY68558.1 ABY68555.1 JTDY01000556 KOB76688.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
C0J8H3
H9JH32
C0J8G4
H9JH34
C0J8G6
H9JH33
+ More
C0J8G9 H9JH30 C0J8H4 C0J8H2 H9J5E5 C0J8H9 C0J8G8 Q25495 Q25494 A0A3G1T1R3 Q25496 G9F9J2 A0A3G1T1Q1 Q7JQ70 Q25500 Q25526 P14754 Q25501 Q25491 Q25492 Q25499 Q25497 Q25498 Q25493 A0A385XRT7 A0A385XRR7 A0A1B6KER4 A0A1B6K9M0 G9F9J3 A0A1B6LVE8 A0A212EIQ3 A0A1B6MQA5 W5ZZB8 A0A1B6L1S1 A0A1B6L303 A0A1B6IT90 W8C425 A0A0A0U6U7 A0A2W1BH42 F5B4G8 H9J5E6 A0A1B6IUV9 V5WCC7 A0A1B6HES2 A0A1B6J4R9 B0FZ67 A0A0P4WD07 A0A0P4W1G3 A0A1B6I5M5 A0A0N9EI66 A0A0P4W9M9 A0A0P4W7J0 A0A0N7ZBP8 A0A0A1X6Z0 A0A0A1WVW1 B4XHA1 A0A0A1XI57 A0A2P8YBQ0 B4XHA0 A0A0R1DLU4 A0A0R1DT85 A0A0R1DR25 A0A0P4WLT4 A0A0R1DSU2 A0A0R1DSX8 B4XH99 A0A1B6FM52 A0A059VDA6 B0FZ64 B0FZ62 B0FZ66 B0FZ63 B0FZ65 B0FZ68 A0A0L7LMC8
C0J8G9 H9JH30 C0J8H4 C0J8H2 H9J5E5 C0J8H9 C0J8G8 Q25495 Q25494 A0A3G1T1R3 Q25496 G9F9J2 A0A3G1T1Q1 Q7JQ70 Q25500 Q25526 P14754 Q25501 Q25491 Q25492 Q25499 Q25497 Q25498 Q25493 A0A385XRT7 A0A385XRR7 A0A1B6KER4 A0A1B6K9M0 G9F9J3 A0A1B6LVE8 A0A212EIQ3 A0A1B6MQA5 W5ZZB8 A0A1B6L1S1 A0A1B6L303 A0A1B6IT90 W8C425 A0A0A0U6U7 A0A2W1BH42 F5B4G8 H9J5E6 A0A1B6IUV9 V5WCC7 A0A1B6HES2 A0A1B6J4R9 B0FZ67 A0A0P4WD07 A0A0P4W1G3 A0A1B6I5M5 A0A0N9EI66 A0A0P4W9M9 A0A0P4W7J0 A0A0N7ZBP8 A0A0A1X6Z0 A0A0A1WVW1 B4XHA1 A0A0A1XI57 A0A2P8YBQ0 B4XHA0 A0A0R1DLU4 A0A0R1DT85 A0A0R1DR25 A0A0P4WLT4 A0A0R1DSU2 A0A0R1DSX8 B4XH99 A0A1B6FM52 A0A059VDA6 B0FZ64 B0FZ62 B0FZ66 B0FZ63 B0FZ65 B0FZ68 A0A0L7LMC8
PDB
1SEK
E-value=9.63254e-14,
Score=185
Ontologies
PANTHER
Topology
Subcellular location
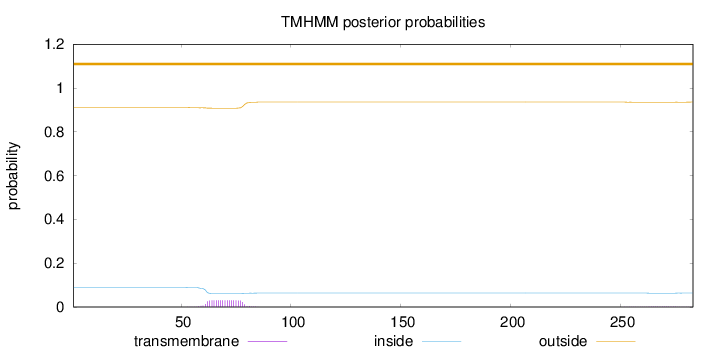
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.59638
Exp number, first 60 AAs:
0.01773
Total prob of N-in:
0.08954
outside
1 - 283
Population Genetic Test Statistics
Pi
254.530769
Theta
180.283287
Tajima's D
2.929023
CLR
0.318927
CSRT
0.975901204939753
Interpretation
Uncertain
