Gene
KWMTBOMO01591
Pre Gene Modal
BGIBMGA008831
Annotation
serine_protease_inhibitor_15_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.3
Sequence
CDS
ATGTTTCGGATCCTGACGCTCTGTTGTGTCCTGACGTTAATGTTGGTTGAAGCTAATAGAGACTTATTAGAAGCTAAAAAACGTTTAGATGCTGCCAACAGTCGAATAATCGGAGAATTTCTATATCAAGGTGCCAAAAGAAACCCTGGTGAAATCTATACGTCTGCACTATTACCTATTTTGGCAGCCGCAGGACAGCTTGCGTTGTTCTGCAGAGAGAAAACGTGTGATCAGCTATTGAACTTCCTAAATTTAAAATATAAAGCAGAGATTCTCGTGCTTTTTCCTATACTAAGGAAGACATTATTCGATGAAAGTCTAATCGGTTCGCCGTCACTGAATTTAAATGTCTACGCTGATGAGGACAACAAGTTCTGTAAGGCGTTCACTCAATCCTACTACAAGACCTTCGAGGGTAAGACCAGGCTTGTGGACTACAGTGACCCGGTGGGAGCGGCTAAGGAGATTAATAATGAGGTAAAAGAGGAAGGCTCAGGAAACTTCAAGAGCTTAATTTCTAGTAAGATGATAAAGGAAGGAGCCGGTCTCAATTTTGTAAGCACCTACGAGCAGACTGTTATATACGACGATCAATTTATATTCAAGAAAAACGTAACGATAAAATTCAAATCCGGCCGCAAGGTGACCAACGTCCCGGGCATCGCCGGGGAAGGTATCGTGAAATATGCCGACCTAAAAGATTTTGACGCCAAGAGCTATGAACTGGTCGGCGAAGGTGAAGTAAACTCGTATCTCATAGTTGTTCCGAACAAACCTGATGGACTTCTCGGATTATTGGATAAATTACGTAGTCCTACAGCTTTTGGAGATATTCTTGAACGCGGAGAAGAAGTTTGTGCGAAAATATCGATCTTATACAGGGACTTTATTGCTAAGATCGATTTAGGGAAGTCTGCCCGAGAGTCAAATTACTTGGAGGGTGCTTTCCAAGAGGGAGGTCTTCAGCTCCGAGGTGTAAGTCAAAAGTGCAACAAAACGTACATCTCGTCAGTTCTACACGAAGTTTCCTTGAAATTAGGATTTGGTAATGGATACAAAGATGCGATGCGGATCGGCTGGTGTCCCGTCAGCGAGATACGGCAAACTGACGCCTCGATATGTAAGAGCGAGGGAGTATCGAGAGAGACACAGTGCAGGGCCCGCCTATAGTGCACACTAACCGACTGGGTGGGCCGCGACCGCGGATGCCTCACCTTCCGGCTGACGCAGATGTTGACCGGCCATGGGTGTTTTGGCCGGTACCTCTTCAAGATAGCCGGAAGGGAACCGACGGCGCAGTGCCACCACTGCGCGGATCGCGACGAAGAAGACACAGCGGAACATACATTGGCGCGTTGCTCTGGTTTCGATGAGCAACGCGCCGCCCTCGTCGCGGTCATAGGAGAGGACCTCTCGCTGCCGCGCGTCGTGGCTACGATGCTCGGCAGCGACGCGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAGCGAGAGAGGGAGAGCTCTTCCCTTTCCGCACCGATCCGTCGCCGTCGAGCCGGGGGCCGGAGGCGGGAATACGCCCACTTATTAGAAGCTAAGAAACGTTTAGATGCTGCCAACAATCGAATAACTGGAGAATTTCTATATCAAGGAGCAAAAAGAAACCCTGGTGAAATCTATACGTCTGCACTATATCCTATTTTGGTAGCCGCGGGACAGCTCGCATTATTCTGCAAGGAGACAACGTGTGATCATCTACTTAAGTTCCTAAATTTGAAAGACAAAGCAGAGATTCTCGCGCTTTTTCCTACAATAAGGAGGATATTTTTCGACGAACCCCTAATCGGTTTGCCGTCACTGAATTTAAATGTCTACGCTGATAAGGACAACAAGTTCTGTAAGGCGTTCACTCAATCCTACTACAAGACCTTCGGGGGTAAGACCAGGCTTGTGGACTACAGTGACCCGGTGGGAGCGGCTAAAGAGATTAATAATGAGGTAAAAGAGGAAGGCTCAGGAAGATTCAAGAGTTTAATTTCTAGTAAGATGATAAAGGAAGGAGCCGGTCTCAATTTCGTAAGCACCTACGAGCAGACTATCATATACGACGATCAATTTATATTCAAGAAAAACGTAACGATAAAATTCAAATCCGGCCGCAAGGTGACCAACGTCCCGGGAATCGCCGGGGAAGGTATCGTGAAATATGCCGACCTAAAAGATTTTGACGCCAAGAGCTATGAACTGGTTGGCGAACGTGGATTATGTTCATATCTCATACTTGTTCCGAACAAACCAGATGGACTTCTTGGATTATTGGACAAATTACGTAGTCCTACAGCTTTTGGAGAAATTCTTGAACGCGGAGAAGAAGTTTGTGCAAAAATATCGATCTTATACAGAGACTTAATCGCTAAGGTTGATTTAGGAAAATCTGCCCGAGAGTCAAATTACTTGGAGGGTGCTTTCCAAGAGGGAGGTCTCCAGCTTCGGGGTGTAAGTCAAAATTGCAACAATACATACATCTCCTCGGTTCTACACGAGGTTTCCTTGAAATCAAGGTTTAGTAATGGATACAAAGCTAGTAAATGTTCCAAATTCGACGTGGAGCTCGTTACCAACCGTCCTCACTTGTACTTCGTGACGTATACTAAGAAGGTTAGCCCGATCCGCACGCCCGTAGTAGGGGTGGTCGTCGACCTAACTACCAACTAA
Protein
MFRILTLCCVLTLMLVEANRDLLEAKKRLDAANSRIIGEFLYQGAKRNPGEIYTSALLPILAAAGQLALFCREKTCDQLLNFLNLKYKAEILVLFPILRKTLFDESLIGSPSLNLNVYADEDNKFCKAFTQSYYKTFEGKTRLVDYSDPVGAAKEINNEVKEEGSGNFKSLISSKMIKEGAGLNFVSTYEQTVIYDDQFIFKKNVTIKFKSGRKVTNVPGIAGEGIVKYADLKDFDAKSYELVGEGEVNSYLIVVPNKPDGLLGLLDKLRSPTAFGDILERGEEVCAKISILYRDFIAKIDLGKSARESNYLEGAFQEGGLQLRGVSQKCNKTYISSVLHEVSLKLGFGNGYKDAMRIGWCPVSEIRQTDASICKSEGVSRETQCRARLXCTLTDWVGRDRGCLTFRLTQMLTGHGCFGRYLFKIAGREPTAQCHHCADRDEEDTAEHTLARCSGFDEQRAALVAVIGEDLSLPRVVATMLGSDASWKAMLDFCESTISQKEAAERERESSSLSAPIRRRRAGGRRREYAHLLEAKKRLDAANNRITGEFLYQGAKRNPGEIYTSALYPILVAAGQLALFCKETTCDHLLKFLNLKDKAEILALFPTIRRIFFDEPLIGLPSLNLNVYADKDNKFCKAFTQSYYKTFGGKTRLVDYSDPVGAAKEINNEVKEEGSGRFKSLISSKMIKEGAGLNFVSTYEQTIIYDDQFIFKKNVTIKFKSGRKVTNVPGIAGEGIVKYADLKDFDAKSYELVGERGLCSYLILVPNKPDGLLGLLDKLRSPTAFGEILERGEEVCAKISILYRDLIAKVDLGKSARESNYLEGAFQEGGLQLRGVSQNCNNTYISSVLHEVSLKSRFSNGYKASKCSKFDVELVTNRPHLYFVTYTKKVSPIRTPVVGVVVDLTTN
Summary
Uniprot
ProteinModelPortal
PDB
1K9O
E-value=3.55153e-12,
Score=177
Ontologies
GO
Topology
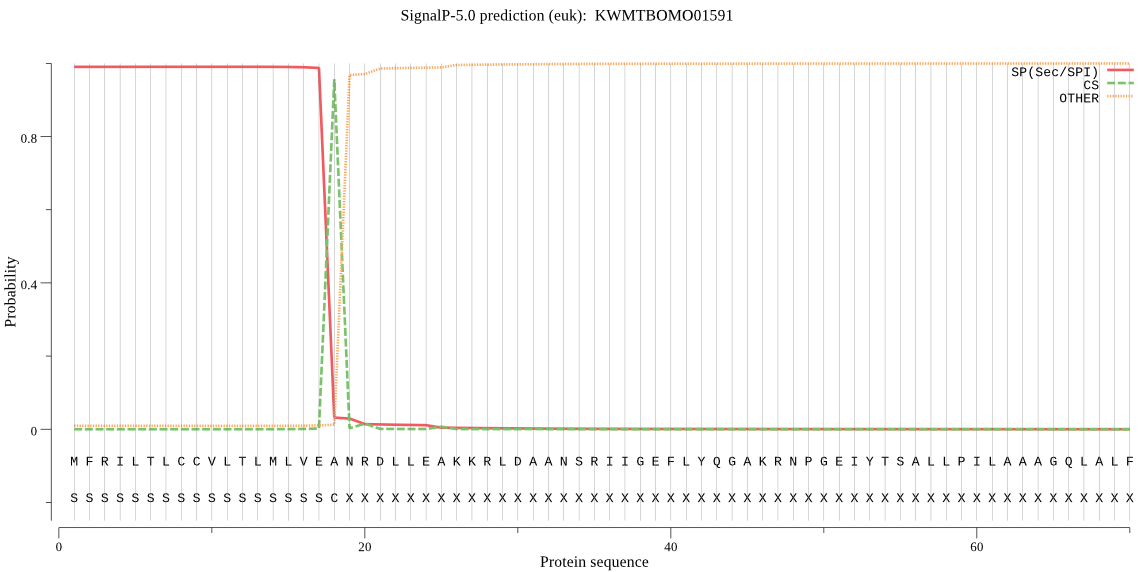
SignalP
Position: 1 - 18,
Likelihood: 0.989301
Length:
907
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.613619999999999
Exp number, first 60 AAs:
0.31024
Total prob of N-in:
0.02847
outside
1 - 907

Population Genetic Test Statistics
Pi
283.392106
Theta
176.589784
Tajima's D
2.14313
CLR
0.172614
CSRT
0.904304784760762
Interpretation
Uncertain
