Gene
KWMTBOMO01582 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008815
Annotation
juvenile_hormone_diol_kinase_[Manduca_sexta]
Location in the cell
Cytoplasmic Reliability : 1.342
Sequence
CDS
ATGGTGTCAGATTTTAGAAAGAAGAAGTTGTTGCACGTTTTCACCGCTTTCTTCGACACCAATGGCAGCGGAACCATTGATAAAAAGGACTTTGAACTTGCCATTGAAAGGATATCCAAATCGAGAGGCTGGAGTGCTGGAGATGCTCAGTACAAAGAAGTTCAAGACACGCTTTTGAAGGTCTGGGATGGGTTGAGCAGCGCTGACACCGATAACGACGGCCAAGTCTCCAAAGAAGAGTGGATTTCCCTCTGGGAGAAGTTTTCCAGCAGCCCCAGTGACTGGCAGAATTTATACTGCAAGTTTATATTTCAACTTGAGGATGCTAGTAATGACGGGTCCATCGACAGCGAGGAATTCTCTTCGGTCTACGCATCTTTTGGTCTAGACAAGGCTGAAGCTGCATCAGCTTTTCAGAAGCTATCCAAAGGCAAGTCATCTGTCTCCTTCGCTGAATTCCAGGAATTGTTCAAAGAATATTTCGCTTCTGAGGATGTGAACGCCCCTGGAAACTTTGTCTTCGGGAAGACCAGCTTCTGA
Protein
MVSDFRKKKLLHVFTAFFDTNGSGTIDKKDFELAIERISKSRGWSAGDAQYKEVQDTLLKVWDGLSSADTDNDGQVSKEEWISLWEKFSSSPSDWQNLYCKFIFQLEDASNDGSIDSEEFSSVYASFGLDKAEAASAFQKLSKGKSSVSFAEFQELFKEYFASEDVNAPGNFVFGKTSF
Summary
Uniprot
H9JH18
Q8T8F5
A0A2A4ITA6
A0A0K8SEY9
A0A2W1BXG2
A0A194Q2G0
+ More
A0A0N1IF43 A0A0L7LAQ1 A0A0H4D9H0 A0A212F2G8 A0A2H1VWL1 A0A2H1WDL9 A0A2W1BQY1 A0A2A4IV08 A0A1Z2RRH9 A0A0L7KJ71 E0ADH1 M4MFD5 Q6F457 A0A194Q368 G5CV04 A0A0N0PC97 H9JA99 A0A0K8U6K4 A0A2W1BNX4 A0A0A1X408 G9G8G8 A0A023EIK2 A0A212F8B7 A0A194Q8J7 A0A1S4FRE5 A0A1Q3FQ01 B0WFD9 W8BI89 Q16RX5 A0A034VMI5 B4LXQ8 A0A194Q2F8 B4K5G7 Q6URH4 H9JH17 A0A182NE20 A0A182F8R2 A0A182YEQ4 A0A0N1INE4 D6W8H7 A0A182Q5H4 A0A1A9WGN2 A0A336MZA6 T1PGT8 Q29B52 B4GZ32 O16158 A0A084W7Z8 B3M2M3 B4JRD8 A0A1L8DBT2 A0A1I8QAP0 A0A1W4UKS1 Q7Q7S2 A0A182IB05 V5G4W4 A0A182RE79 B4QX10 B4PMZ0 B4IBT6 B3P0I7 A0A1A9ZZ27 A0A0F6YNR3 A0A3B0KA82 A0A1W7R574 B4NF47 Q16RX4 U4U4F5 J3JX69 A0A1A9YSX4 A0A1B0B2M3 A0A182J1T9 A0A1J1I1W7 A0A1Y1LJT5 E2BCD9 A0A0M4F458 H9JH16 R4V1R5 A0A182VPW7 E2ADZ5 A0A2S1TNT2 A0A0L0BY56 A0A1B6C3R8 A0A1B0FEJ6 A0A0N0PAU1 A0A0N0PB40 A0A026W3T2 A0A1W4XCU0 B0WFE0 A0A3L8DTH4 A0A2S2R736 A0A088AE24 V9I7D4 A0A1L8DBI6
A0A0N1IF43 A0A0L7LAQ1 A0A0H4D9H0 A0A212F2G8 A0A2H1VWL1 A0A2H1WDL9 A0A2W1BQY1 A0A2A4IV08 A0A1Z2RRH9 A0A0L7KJ71 E0ADH1 M4MFD5 Q6F457 A0A194Q368 G5CV04 A0A0N0PC97 H9JA99 A0A0K8U6K4 A0A2W1BNX4 A0A0A1X408 G9G8G8 A0A023EIK2 A0A212F8B7 A0A194Q8J7 A0A1S4FRE5 A0A1Q3FQ01 B0WFD9 W8BI89 Q16RX5 A0A034VMI5 B4LXQ8 A0A194Q2F8 B4K5G7 Q6URH4 H9JH17 A0A182NE20 A0A182F8R2 A0A182YEQ4 A0A0N1INE4 D6W8H7 A0A182Q5H4 A0A1A9WGN2 A0A336MZA6 T1PGT8 Q29B52 B4GZ32 O16158 A0A084W7Z8 B3M2M3 B4JRD8 A0A1L8DBT2 A0A1I8QAP0 A0A1W4UKS1 Q7Q7S2 A0A182IB05 V5G4W4 A0A182RE79 B4QX10 B4PMZ0 B4IBT6 B3P0I7 A0A1A9ZZ27 A0A0F6YNR3 A0A3B0KA82 A0A1W7R574 B4NF47 Q16RX4 U4U4F5 J3JX69 A0A1A9YSX4 A0A1B0B2M3 A0A182J1T9 A0A1J1I1W7 A0A1Y1LJT5 E2BCD9 A0A0M4F458 H9JH16 R4V1R5 A0A182VPW7 E2ADZ5 A0A2S1TNT2 A0A0L0BY56 A0A1B6C3R8 A0A1B0FEJ6 A0A0N0PAU1 A0A0N0PB40 A0A026W3T2 A0A1W4XCU0 B0WFE0 A0A3L8DTH4 A0A2S2R736 A0A088AE24 V9I7D4 A0A1L8DBI6
Pubmed
19121390
11909870
28756777
26354079
26227816
25959665
+ More
22118469 28605547 25830018 24945155 24495485 17510324 25348373 17994087 16203205 25244985 18362917 19820115 25315136 15632085 9443378 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 12364791 14747013 17210077 17550304 23537049 22516182 28004739 20798317 29727440 26108605 24508170 30249741
22118469 28605547 25830018 24945155 24495485 17510324 25348373 17994087 16203205 25244985 18362917 19820115 25315136 15632085 9443378 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 12364791 14747013 17210077 17550304 23537049 22516182 28004739 20798317 29727440 26108605 24508170 30249741
EMBL
BABH01037604
AJ430670
CAD23378.1
NWSH01006845
PCG63217.1
GBRD01014476
+ More
JAG51350.1 KZ149928 PZC77500.1 KQ459562 KPI99731.1 KQ461158 KPJ08058.1 JTDY01001913 KOB72552.1 KP192493 AKN89772.1 AGBW02010752 OWR47916.1 ODYU01004874 SOQ45193.1 ODYU01007837 SOQ50962.1 PZC77499.1 PCG63218.1 KY938810 ASA46451.1 JTDY01009505 KOB63382.1 HM626514 ADK94879.2 KC127671 AGG56523.1 AB180433 BAD26677.1 KQ459579 KPI99449.1 JN410823 AEP25399.1 KQ460650 KPJ13114.1 BABH01008872 BABH01008873 GDHF01030103 GDHF01016478 GDHF01008965 JAI22211.1 JAI35836.1 JAI43349.1 KZ150051 PZC74420.1 GBXI01008480 JAD05812.1 JN256054 AEN03041.1 GAPW01004822 JAC08776.1 AGBW02009771 OWR49958.1 KPI99730.1 GFDL01005365 JAV29680.1 DS231916 EDS26156.1 GAMC01013584 JAB92971.1 CH477693 EAT37200.1 GAKP01015927 GAKP01015924 JAC43025.1 CH940650 EDW67867.1 KPI99726.1 CH933806 EDW16193.1 AY363308 AAQ63486.1 KPJ08053.1 KQ971312 EEZ98300.2 AXCN02002212 UFQS01002765 UFQT01002765 SSX14626.1 SSX34023.1 KA646045 KA647919 AFP62548.1 CM000070 EAL27147.1 CH479198 EDW28050.1 AF014952 AF093240 AE014297 AY089680 AAB67805.1 AAC62484.1 AAF55336.1 AAL90418.1 ATLV01021328 ATLV01021329 ATLV01021330 KE525316 KFB46342.1 CH902617 EDV42344.1 CH916373 EDV94328.1 GFDF01010175 JAV03909.1 AAAB01008952 EAA10596.3 APCN01006585 GALX01003381 JAB65085.1 CM000364 EDX12664.1 CM000160 EDW97002.1 CH480827 EDW44844.1 CH954181 EDV48813.1 KP295467 AKF11872.1 OUUW01000005 SPP80458.1 GEHC01001375 JAV46270.1 CH964251 EDW83422.1 EAT37201.1 KB632098 ERL88789.1 BT127837 AEE62799.1 JXJN01007683 CVRI01000026 CRK92369.1 GEZM01053938 JAV73903.1 GL447257 EFN86675.1 CP012526 ALC46120.1 KC740808 AGM32632.1 GL438827 EFN68228.1 MF189034 AWI63388.1 JRES01001160 KNC24955.1 GEDC01029418 JAS07880.1 CCAG010014864 KPJ08056.1 KPJ08057.1 KK107453 EZA50658.1 EDS26157.1 QOIP01000004 RLU23750.1 GGMS01016307 MBY85510.1 JR035114 JR035115 JR035120 JR035121 JR035134 JR035137 JR035138 JR035141 JR035145 AEY56235.1 AEY56236.1 AEY56237.1 AEY56238.1 AEY56251.1 AEY56252.1 AEY56253.1 AEY56254.1 AEY56258.1 GFDF01010258 JAV03826.1
JAG51350.1 KZ149928 PZC77500.1 KQ459562 KPI99731.1 KQ461158 KPJ08058.1 JTDY01001913 KOB72552.1 KP192493 AKN89772.1 AGBW02010752 OWR47916.1 ODYU01004874 SOQ45193.1 ODYU01007837 SOQ50962.1 PZC77499.1 PCG63218.1 KY938810 ASA46451.1 JTDY01009505 KOB63382.1 HM626514 ADK94879.2 KC127671 AGG56523.1 AB180433 BAD26677.1 KQ459579 KPI99449.1 JN410823 AEP25399.1 KQ460650 KPJ13114.1 BABH01008872 BABH01008873 GDHF01030103 GDHF01016478 GDHF01008965 JAI22211.1 JAI35836.1 JAI43349.1 KZ150051 PZC74420.1 GBXI01008480 JAD05812.1 JN256054 AEN03041.1 GAPW01004822 JAC08776.1 AGBW02009771 OWR49958.1 KPI99730.1 GFDL01005365 JAV29680.1 DS231916 EDS26156.1 GAMC01013584 JAB92971.1 CH477693 EAT37200.1 GAKP01015927 GAKP01015924 JAC43025.1 CH940650 EDW67867.1 KPI99726.1 CH933806 EDW16193.1 AY363308 AAQ63486.1 KPJ08053.1 KQ971312 EEZ98300.2 AXCN02002212 UFQS01002765 UFQT01002765 SSX14626.1 SSX34023.1 KA646045 KA647919 AFP62548.1 CM000070 EAL27147.1 CH479198 EDW28050.1 AF014952 AF093240 AE014297 AY089680 AAB67805.1 AAC62484.1 AAF55336.1 AAL90418.1 ATLV01021328 ATLV01021329 ATLV01021330 KE525316 KFB46342.1 CH902617 EDV42344.1 CH916373 EDV94328.1 GFDF01010175 JAV03909.1 AAAB01008952 EAA10596.3 APCN01006585 GALX01003381 JAB65085.1 CM000364 EDX12664.1 CM000160 EDW97002.1 CH480827 EDW44844.1 CH954181 EDV48813.1 KP295467 AKF11872.1 OUUW01000005 SPP80458.1 GEHC01001375 JAV46270.1 CH964251 EDW83422.1 EAT37201.1 KB632098 ERL88789.1 BT127837 AEE62799.1 JXJN01007683 CVRI01000026 CRK92369.1 GEZM01053938 JAV73903.1 GL447257 EFN86675.1 CP012526 ALC46120.1 KC740808 AGM32632.1 GL438827 EFN68228.1 MF189034 AWI63388.1 JRES01001160 KNC24955.1 GEDC01029418 JAS07880.1 CCAG010014864 KPJ08056.1 KPJ08057.1 KK107453 EZA50658.1 EDS26157.1 QOIP01000004 RLU23750.1 GGMS01016307 MBY85510.1 JR035114 JR035115 JR035120 JR035121 JR035134 JR035137 JR035138 JR035141 JR035145 AEY56235.1 AEY56236.1 AEY56237.1 AEY56238.1 AEY56251.1 AEY56252.1 AEY56253.1 AEY56254.1 AEY56258.1 GFDF01010258 JAV03826.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000002320 UP000008820 UP000008792 UP000009192 UP000075884 UP000069272 UP000076408 UP000007266 UP000075886 UP000091820 UP000095301 UP000001819 UP000008744 UP000000803 UP000030765 UP000007801 UP000001070 UP000095300 UP000192221 UP000007062 UP000075840 UP000075900 UP000000304 UP000002282 UP000001292 UP000008711 UP000092445 UP000268350 UP000007798 UP000030742 UP000092443 UP000092460 UP000075880 UP000183832 UP000008237 UP000092553 UP000075920 UP000000311 UP000037069 UP000092444 UP000053097 UP000192223 UP000279307 UP000005203
UP000002320 UP000008820 UP000008792 UP000009192 UP000075884 UP000069272 UP000076408 UP000007266 UP000075886 UP000091820 UP000095301 UP000001819 UP000008744 UP000000803 UP000030765 UP000007801 UP000001070 UP000095300 UP000192221 UP000007062 UP000075840 UP000075900 UP000000304 UP000002282 UP000001292 UP000008711 UP000092445 UP000268350 UP000007798 UP000030742 UP000092443 UP000092460 UP000075880 UP000183832 UP000008237 UP000092553 UP000075920 UP000000311 UP000037069 UP000092444 UP000053097 UP000192223 UP000279307 UP000005203
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
H9JH18
Q8T8F5
A0A2A4ITA6
A0A0K8SEY9
A0A2W1BXG2
A0A194Q2G0
+ More
A0A0N1IF43 A0A0L7LAQ1 A0A0H4D9H0 A0A212F2G8 A0A2H1VWL1 A0A2H1WDL9 A0A2W1BQY1 A0A2A4IV08 A0A1Z2RRH9 A0A0L7KJ71 E0ADH1 M4MFD5 Q6F457 A0A194Q368 G5CV04 A0A0N0PC97 H9JA99 A0A0K8U6K4 A0A2W1BNX4 A0A0A1X408 G9G8G8 A0A023EIK2 A0A212F8B7 A0A194Q8J7 A0A1S4FRE5 A0A1Q3FQ01 B0WFD9 W8BI89 Q16RX5 A0A034VMI5 B4LXQ8 A0A194Q2F8 B4K5G7 Q6URH4 H9JH17 A0A182NE20 A0A182F8R2 A0A182YEQ4 A0A0N1INE4 D6W8H7 A0A182Q5H4 A0A1A9WGN2 A0A336MZA6 T1PGT8 Q29B52 B4GZ32 O16158 A0A084W7Z8 B3M2M3 B4JRD8 A0A1L8DBT2 A0A1I8QAP0 A0A1W4UKS1 Q7Q7S2 A0A182IB05 V5G4W4 A0A182RE79 B4QX10 B4PMZ0 B4IBT6 B3P0I7 A0A1A9ZZ27 A0A0F6YNR3 A0A3B0KA82 A0A1W7R574 B4NF47 Q16RX4 U4U4F5 J3JX69 A0A1A9YSX4 A0A1B0B2M3 A0A182J1T9 A0A1J1I1W7 A0A1Y1LJT5 E2BCD9 A0A0M4F458 H9JH16 R4V1R5 A0A182VPW7 E2ADZ5 A0A2S1TNT2 A0A0L0BY56 A0A1B6C3R8 A0A1B0FEJ6 A0A0N0PAU1 A0A0N0PB40 A0A026W3T2 A0A1W4XCU0 B0WFE0 A0A3L8DTH4 A0A2S2R736 A0A088AE24 V9I7D4 A0A1L8DBI6
A0A0N1IF43 A0A0L7LAQ1 A0A0H4D9H0 A0A212F2G8 A0A2H1VWL1 A0A2H1WDL9 A0A2W1BQY1 A0A2A4IV08 A0A1Z2RRH9 A0A0L7KJ71 E0ADH1 M4MFD5 Q6F457 A0A194Q368 G5CV04 A0A0N0PC97 H9JA99 A0A0K8U6K4 A0A2W1BNX4 A0A0A1X408 G9G8G8 A0A023EIK2 A0A212F8B7 A0A194Q8J7 A0A1S4FRE5 A0A1Q3FQ01 B0WFD9 W8BI89 Q16RX5 A0A034VMI5 B4LXQ8 A0A194Q2F8 B4K5G7 Q6URH4 H9JH17 A0A182NE20 A0A182F8R2 A0A182YEQ4 A0A0N1INE4 D6W8H7 A0A182Q5H4 A0A1A9WGN2 A0A336MZA6 T1PGT8 Q29B52 B4GZ32 O16158 A0A084W7Z8 B3M2M3 B4JRD8 A0A1L8DBT2 A0A1I8QAP0 A0A1W4UKS1 Q7Q7S2 A0A182IB05 V5G4W4 A0A182RE79 B4QX10 B4PMZ0 B4IBT6 B3P0I7 A0A1A9ZZ27 A0A0F6YNR3 A0A3B0KA82 A0A1W7R574 B4NF47 Q16RX4 U4U4F5 J3JX69 A0A1A9YSX4 A0A1B0B2M3 A0A182J1T9 A0A1J1I1W7 A0A1Y1LJT5 E2BCD9 A0A0M4F458 H9JH16 R4V1R5 A0A182VPW7 E2ADZ5 A0A2S1TNT2 A0A0L0BY56 A0A1B6C3R8 A0A1B0FEJ6 A0A0N0PAU1 A0A0N0PB40 A0A026W3T2 A0A1W4XCU0 B0WFE0 A0A3L8DTH4 A0A2S2R736 A0A088AE24 V9I7D4 A0A1L8DBI6
PDB
4NDC
E-value=1.23479e-30,
Score=327
Ontologies
GO
Topology
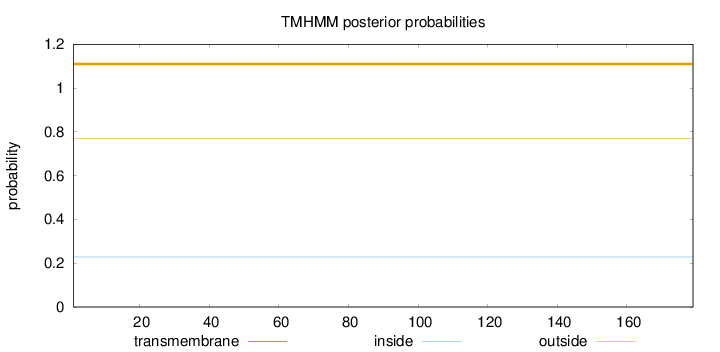
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00106
Exp number, first 60 AAs:
0.00106
Total prob of N-in:
0.22891
outside
1 - 179
Population Genetic Test Statistics
Pi
269.076931
Theta
209.3118
Tajima's D
0.898915
CLR
0
CSRT
0.628368581570921
Interpretation
Uncertain
