Gene
KWMTBOMO01579 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008812
Annotation
putative_vesicle_associated_protein_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.394 Nuclear Reliability : 1.098
Sequence
CDS
ATGAAACTGAAGGAGTTAAAGAGGACTGTGAACGTGAGCTGGTCCCCGGCTGATCATCACCCGGTAACGCTTGTTGCCGGCTCGGCGGCCCAGCAAGTTGACGCGTCTTTCAGCTCAAGTTCATACTTAGAACTGTATTCGTTGAATTTAGAAGACCCTAGTTATGATTTGAAACAGATATCCAGTTTGCAAACACAACATAAATTCCATAAGATAGAATGGTCAGGTGCCGGAATTATCATAGGAGGCTGTGATGGTGGATTATTAGAGTTTTACAGTGCTGAAAAACTGTTGAACAATGCCAGTGATGCTTTACTTGGAAGCAGTACGAAGCATAGTGGACATGTCTCCGCTTTGGATGTTAATCCATACCAGAAGAATCTGTTGGCCTCAGGAGCGTCAGAAAGTGAGATATTCATATGGGACCTGAACAACACAAGTCAACCAATGGCTCCTGGCACTAGGAGCCAACCATATGATCATGTGCAGGGCTTGGCTTGGAATCAGCAGGTACAACATATTCTAGGTTCAACGTTCGCGACAAGGTGTGTCGTCTGGGATCTCAGAAAAAATGAACCCATCATGAAATTAAGCGACTCCCAGTCCCGCAGCCGGTGGCGCGCCGTGAGCTGGCACCCCGGGGTCGCCACGCAGCTCTGCCTTGCCTCCGAGGACGACCAGGCGCCCGTCATACAGCTCTGGGACCTCAGGCTGGCGGCGTCGCCGGTGGTGTCGCTGGAGGGGCACGGCAAGGGCGCGCTGTCCATGTCGTGGTGCGCGCAGGACGCCGACCTGCTGCTCTCCGCCGGGAAGGACGGCCGGGTGCTGTGCTGGAACCCGAATACCACTAAACCGGGTGGCGAGGTGGTGGCGGAGTGCGCGGGGTCGGCGCAGTGGGTGTTCGAAGTGCGCTGGTGTCCGCGCGTGCCCGCCCTGTTCGCCGCCGCTGCTTTTGACCAGCGCCTGGCTGTGCACTCGGTGCTTGGAGGCGCCGCAGCTGATACGAGTAGTCAGAGTCAGAGCAACATCATGGACTCGTTCGGCGCCGAGTCGTTCAGCCAGCTGCCGGCCGTGCGGGGCGCGCAGGGGGCGGGGGGCGGCGCGGGGGCAGGCGCGGGGGCCGGGCGGGGGGCGCTGGCGGCGCCGCCACGTTGGATGAAGCGCCCGGTGCGGGCCAACTTCGGGTTTGGTGGTAAACTGGTGACGTTTGAGAAGTGTCCGCAACAAGAGGGCGGCGCTCAGAGACTGGTGCACATTAGCCAGGTGGTGAGCGATCCGGAGATAGTGGAGCTGGCAGCGGAGTTGGACGCGGTGCTAGGTTCGAGTCCCAGCCAAGACCCCGAGGCCAACCAGCGGCTCGCCGAGTACTGCCGACAGAGGGGTGACGCGGCAACCGACCAGAACGAGCGCTACACGTGGTTCTTCCTGCGAGCCAACTTCCTGCCTTCCTTCCGGTCCGAAGTACTCAATCTACTTGGGTTCAGACAAGACGAGATCCCGTCGAAGTTCAAAACCCTGGACATGTCCGGAGACGAAGCGCCGCCCGACCCAGCCCTGCCGCACGTGGAGGGGCTGTCGCTGCTGGACCGCAAGCTGGCGGACGTGGAGTCGGAGCCCGCGGTCAGCGACGTGCACATACCCAACGGCGACGATGAGACGAGCATGATATGCCGCGCGCTGGTGTGCGGCAACCTGGAGGAGGCCGTGGAGCTGTGCCTGGCGGCGGAGCGGGTGGCCGACGCGCTCGTCATCGCCTCGCTCGGCAGTCAAGAGCTAGTGTACAAAGTGCAGAAGTACCACCTGAACCGCACGTGCGCGGAGCCGGTGTCGGCGCTGGCGGACGCGCTGCTGGCGGCGCGGTGGGGCGCGCTGCTGGCACGGACCGGCGCGCGCGCCTGGCGCAGCGTGCTGGCCGCGCTCGTCACGCACTGCGACACCGACGCGCTGCCGCACTACTGTGAAACTTTAGGGGATAAGTTATCCAAGGAGTCAGATCCAGCTCTGGTGCAGGCCGCCACGATCTGCTACCTGTGCGCCATGAGCCCCGAGCGGCTGGCGCGGGTGTGGGGGGCGCTCCCCCCGCGCCACGCCGCGCCCCCCGCGCTCCTCGCGCACCGCGCCGCCGCCCTGCGACACGCGCAGCTACAGTCCGGCGGTAAACTGGAGACAATACTGAACGAGTACGCGCTGAACCTCGCCTCACAAGGCTGTTTGCAGAGCGCCCTGCTGGCCGCGCAGGACTCCGGGTCCGACGACCTCAAGCAGCGGCTGCAGAGGGCGCTGGGAATCACGCGAGACCCCGCGCACAAGGCCCGGGCGCCGGCACACGGAGGGGCGCGCGGCTACGGGCGGCGCGATGACGTCACGGCGCACTACAACTCCGCCGTCCCGCCGCAGTACGATCCGTCGCCATGGCAGCAGTCCACAGTCTCAGCGCACCAGTACTCGCAGCCGCCTCTTGCACCCCTCGCTCCGCAGCCTCTCGCCCCCCAAACGTTCGCCCCCCAGCCCCTAGCCCCACAGCCCCTCGTCCCCCAGCCCCCCGCCCCGCAGCCGCCCCCGCCCGCGGCCCCGCCCCGCCCCGGAAGCGTCGGGCCTACAGCTGGCGGCCTCGCGTCGCGCTCCAAGTACCGGGTGGACCCCTCCGTGCAGCCCGCGCCGCCCTGCGCCCCCTACCCCGCCCGCCCCTACACGCCCACACACCCCGCAGTCAACGGCACCTACTACAACCCTCCAGGTGTGGAGACGGGGGTGCCCGTGGTCCCCACCCAGAACAAGCCCGCGGCCCCCGGCTGGAACGACCCGCCGATGCTGTCGTCTAAACCTAAGGCTAAATCGGAGACGCCCGTACCAGCTCAAGCGCCGATAACGCATCCGTTGTTCGGAGTGGAGCCCCCGCAGCACATACCCTTAGCCCCCGCACCCGGGCAGTACCCCGGGATGCAACCGGGGCAGTACCAGGGCTCACAGTTCCCGGCACCACAACCGTCGGTCGGACAATATTCGATTGTACAACAACCAATCGGACAATACCCAGGAGGACAACAACACATTGGACAATATCCGGGGGCACAACAACCGGATGGGCAGTATCCTGGGGCACAACAGCTCGGTGGACAGTACCCCGGGGCACAATACCCGGGCGGGCAATACCAAGATCAGCAATATCAGGCTCAGTATCCCCCACAACCACCTCCACAACAATACACTCCTCAGGTGGAGCCTCAGCCCGCCCCCAAGCCCCCGCTGCCGCCCCCCCTCGCGGCGCTGCAGGACACGTTCGACGGGCTCCGGGAGCAGCTCGCGCGCGCCTCCACCAGCTCGCTCGTGAAACGGAAACTGGAAGATGTACAGAGAAGACTGGAGACCATGTACGACTTGCTGAGGGAGGACCGGCTGTCGGAGAGCTGCGTGTGTGCGCTGACAGAGATGTCTGCGTGTGTGCGCGCGGGGGACGCGGGGGGCGCGCTGCGGGGCGCGGGGGCGCTGGCAGCGGGGGGCGACTTCGCGGCCGTCGCCAGCTTCCTGCCCGGACTCAAGACGCTCTTCCAGCTCGCCGCCGCCGAGCACGCGCCGCGATAG
Protein
MKLKELKRTVNVSWSPADHHPVTLVAGSAAQQVDASFSSSSYLELYSLNLEDPSYDLKQISSLQTQHKFHKIEWSGAGIIIGGCDGGLLEFYSAEKLLNNASDALLGSSTKHSGHVSALDVNPYQKNLLASGASESEIFIWDLNNTSQPMAPGTRSQPYDHVQGLAWNQQVQHILGSTFATRCVVWDLRKNEPIMKLSDSQSRSRWRAVSWHPGVATQLCLASEDDQAPVIQLWDLRLAASPVVSLEGHGKGALSMSWCAQDADLLLSAGKDGRVLCWNPNTTKPGGEVVAECAGSAQWVFEVRWCPRVPALFAAAAFDQRLAVHSVLGGAAADTSSQSQSNIMDSFGAESFSQLPAVRGAQGAGGGAGAGAGAGRGALAAPPRWMKRPVRANFGFGGKLVTFEKCPQQEGGAQRLVHISQVVSDPEIVELAAELDAVLGSSPSQDPEANQRLAEYCRQRGDAATDQNERYTWFFLRANFLPSFRSEVLNLLGFRQDEIPSKFKTLDMSGDEAPPDPALPHVEGLSLLDRKLADVESEPAVSDVHIPNGDDETSMICRALVCGNLEEAVELCLAAERVADALVIASLGSQELVYKVQKYHLNRTCAEPVSALADALLAARWGALLARTGARAWRSVLAALVTHCDTDALPHYCETLGDKLSKESDPALVQAATICYLCAMSPERLARVWGALPPRHAAPPALLAHRAAALRHAQLQSGGKLETILNEYALNLASQGCLQSALLAAQDSGSDDLKQRLQRALGITRDPAHKARAPAHGGARGYGRRDDVTAHYNSAVPPQYDPSPWQQSTVSAHQYSQPPLAPLAPQPLAPQTFAPQPLAPQPLVPQPPAPQPPPPAAPPRPGSVGPTAGGLASRSKYRVDPSVQPAPPCAPYPARPYTPTHPAVNGTYYNPPGVETGVPVVPTQNKPAAPGWNDPPMLSSKPKAKSETPVPAQAPITHPLFGVEPPQHIPLAPAPGQYPGMQPGQYQGSQFPAPQPSVGQYSIVQQPIGQYPGGQQHIGQYPGAQQPDGQYPGAQQLGGQYPGAQYPGGQYQDQQYQAQYPPQPPPQQYTPQVEPQPAPKPPLPPPLAALQDTFDGLREQLARASTSSLVKRKLEDVQRRLETMYDLLREDRLSESCVCALTEMSACVRAGDAGGALRGAGALAAGGDFAAVASFLPGLKTLFQLAAAEHAPR
Summary
Uniprot
A0A2W1BR35
A0A2H1W0C1
A0A1Y1KL12
A0A1Y1KDG9
A0A1Y1KG21
Q17KX8
+ More
A0A1Q3FE24 Q17KX9 A0A1S4EZ06 A0A084W1I8 A0A0P6IUR7 A0A1S4EYZ4 A0A182FU80 A0A182MII7 A0A2M4BA78 A0A182MXT3 A0A2M4A0X8 A0A2M4B9P9 A0A182QF13 A0A2M4A0W0 A0A2M4A0X1 A0A182WEU1 A0A182Y5Z2 A0A182XC07 A0A182VJT7 A0A2M3YZH3 A0A182HNA5 A0A182PKW7 A0A182K048 A0A182GCS8 A0A182RY04 A0A182J4K5 A0A1B0D3A8 A0A147BUB7 A0A131Z2N2 A0A224Z596 L7M955 A0A224Z752 L7M6L4
A0A1Q3FE24 Q17KX9 A0A1S4EZ06 A0A084W1I8 A0A0P6IUR7 A0A1S4EYZ4 A0A182FU80 A0A182MII7 A0A2M4BA78 A0A182MXT3 A0A2M4A0X8 A0A2M4B9P9 A0A182QF13 A0A2M4A0W0 A0A2M4A0X1 A0A182WEU1 A0A182Y5Z2 A0A182XC07 A0A182VJT7 A0A2M3YZH3 A0A182HNA5 A0A182PKW7 A0A182K048 A0A182GCS8 A0A182RY04 A0A182J4K5 A0A1B0D3A8 A0A147BUB7 A0A131Z2N2 A0A224Z596 L7M955 A0A224Z752 L7M6L4
Pubmed
EMBL
KZ149928
PZC77498.1
ODYU01005270
SOQ45954.1
GEZM01086348
JAV59527.1
+ More
GEZM01086346 JAV59529.1 GEZM01086347 JAV59528.1 CH477220 EAT47377.1 GFDL01009240 JAV25805.1 EAT47376.1 ATLV01019349 KE525268 KFB44082.1 GDUN01001109 JAN94810.1 AXCM01001536 AXCM01001537 GGFJ01000796 MBW49937.1 GGFK01001074 MBW34395.1 GGFJ01000591 MBW49732.1 AXCN02002117 GGFK01001061 MBW34382.1 GGFK01001080 MBW34401.1 GGFM01000928 MBW21679.1 APCN01001898 JXUM01054649 JXUM01054650 JXUM01054651 KQ561831 KXJ77404.1 AJVK01023417 AJVK01023418 AJVK01023419 AJVK01023420 GEGO01001106 JAR94298.1 GEDV01004166 JAP84391.1 GFPF01011085 MAA22231.1 GACK01005356 JAA59678.1 GFPF01011084 MAA22230.1 GACK01005359 JAA59675.1
GEZM01086346 JAV59529.1 GEZM01086347 JAV59528.1 CH477220 EAT47377.1 GFDL01009240 JAV25805.1 EAT47376.1 ATLV01019349 KE525268 KFB44082.1 GDUN01001109 JAN94810.1 AXCM01001536 AXCM01001537 GGFJ01000796 MBW49937.1 GGFK01001074 MBW34395.1 GGFJ01000591 MBW49732.1 AXCN02002117 GGFK01001061 MBW34382.1 GGFK01001080 MBW34401.1 GGFM01000928 MBW21679.1 APCN01001898 JXUM01054649 JXUM01054650 JXUM01054651 KQ561831 KXJ77404.1 AJVK01023417 AJVK01023418 AJVK01023419 AJVK01023420 GEGO01001106 JAR94298.1 GEDV01004166 JAP84391.1 GFPF01011085 MAA22231.1 GACK01005356 JAA59678.1 GFPF01011084 MAA22230.1 GACK01005359 JAA59675.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BR35
A0A2H1W0C1
A0A1Y1KL12
A0A1Y1KDG9
A0A1Y1KG21
Q17KX8
+ More
A0A1Q3FE24 Q17KX9 A0A1S4EZ06 A0A084W1I8 A0A0P6IUR7 A0A1S4EYZ4 A0A182FU80 A0A182MII7 A0A2M4BA78 A0A182MXT3 A0A2M4A0X8 A0A2M4B9P9 A0A182QF13 A0A2M4A0W0 A0A2M4A0X1 A0A182WEU1 A0A182Y5Z2 A0A182XC07 A0A182VJT7 A0A2M3YZH3 A0A182HNA5 A0A182PKW7 A0A182K048 A0A182GCS8 A0A182RY04 A0A182J4K5 A0A1B0D3A8 A0A147BUB7 A0A131Z2N2 A0A224Z596 L7M955 A0A224Z752 L7M6L4
A0A1Q3FE24 Q17KX9 A0A1S4EZ06 A0A084W1I8 A0A0P6IUR7 A0A1S4EYZ4 A0A182FU80 A0A182MII7 A0A2M4BA78 A0A182MXT3 A0A2M4A0X8 A0A2M4B9P9 A0A182QF13 A0A2M4A0W0 A0A2M4A0X1 A0A182WEU1 A0A182Y5Z2 A0A182XC07 A0A182VJT7 A0A2M3YZH3 A0A182HNA5 A0A182PKW7 A0A182K048 A0A182GCS8 A0A182RY04 A0A182J4K5 A0A1B0D3A8 A0A147BUB7 A0A131Z2N2 A0A224Z596 L7M955 A0A224Z752 L7M6L4
PDB
2PM9
E-value=1.08481e-45,
Score=467
Ontologies
GO
PANTHER
Topology
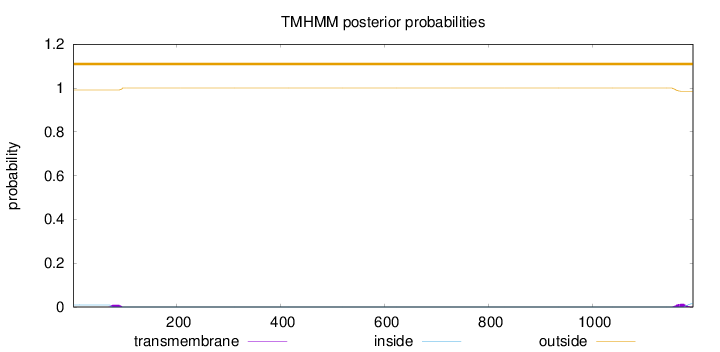
Length:
1193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51546
Exp number, first 60 AAs:
0.00044
Total prob of N-in:
0.00949
outside
1 - 1193
Population Genetic Test Statistics
Pi
267.901694
Theta
172.523242
Tajima's D
1.526255
CLR
0.14935
CSRT
0.789210539473026
Interpretation
Uncertain
