Gene
KWMTBOMO01573
Annotation
PREDICTED:_protein_aveugle_[Amyelois_transitella]
Full name
Protein aveugle
Location in the cell
Mitochondrial Reliability : 1.333 Nuclear Reliability : 1.884
Sequence
CDS
ATGGTGGAAGAACCGAATCTTAATTCAAGTAAACCCAAAGCGAAAACGACCCGTCCCAAAGCTATATATTTATGGACAGAAGCCGACGTCCAAAAGTGGCTTCGACGCCACTGTAGTGATTATTATCATATGTACTGGGAGAGTTTTCACGAGCATGACATAACAGGTAGAGCTCTAGTCCGTATCAATGACAACACATTACTTCGAATGGGTATCAACAACAAAGATCACAGGGAAGCCATATGGAGGGAAATCTTAAAACTTAGACTGAAAGCTGACATAGTCGAGATCCGTGATTTAGAACGTAGGCATAACTATTTTAATTATGACTTGTGA
Protein
MVEEPNLNSSKPKAKTTRPKAIYLWTEADVQKWLRRHCSDYYHMYWESFHEHDITGRALVRINDNTLLRMGINNKDHREAIWREILKLRLKADIVEIRDLERRHNYFNYDL
Summary
Description
Required for normal photoreceptor differentiation between Ras and Raf for EGFR signaling in the eye and for mitogen-activated protein kinase phosphorylation. Probably acts together with Cnk to promote Raf activation, perhaps by recruiting an activating kinase.
Subunit
Interacts with the SAM domain of cnk.
Miscellaneous
'Aveugle' means 'blind' in French.
Keywords
3D-structure
Complete proteome
Cytoplasm
Membrane
Reference proteome
Sensory transduction
Vision
Feature
chain Protein aveugle
Uniprot
A0A2W1BQX2
A0A2H1W6H4
A0A2A4J2T1
A0A1E1WID3
S4PTB1
A0A0N0PAU7
+ More
A0A194Q2G8 A0A2P8ZI11 A0A2J7RQR4 A0A067RD26 A0A0N7Z8Y7 R4G818 A0A0V0G7D0 A0A069DVI1 A0A1B6EFB1 A0A0M8ZVF2 U4UGC3 J3JVP6 A0A1W4WTV8 E2BTE0 R4WQ63 A0A154PEP3 D6WUY3 A0A310SR28 E2ARH1 A0A2A3E0G5 V9IE46 A0A088AK94 A0A0A9Z9B1 A0A1Y1LS78 A0A158NQB1 A0A026WQC8 A0A1B6HSZ2 A0A310SGQ1 A0A151K350 A0A1B6FQ93 K7IQ21 A0A232F8U4 A0A1B6MMI4 E9GS85 A0A195B547 A0A151WGI0 A0A0P5IYV3 A0A0P5PC63 A0A0P4XK11 A0A2R7W450 H9JN51 A0A0T6AX06 A0A0P5FD74 A0A2H8TQD2 A0A2S2PQD1 B4HRN8 A0A0P4WI92 J9JZT8 A0A2S2Q3U3 A0A3Q0JNX5 W8BMX1 A0A1B2AIY1 B3NQT3 B4QFY1 Q8ML92 B4P7N5 B4KNN3 Q16IB6 B4J6L1 A0A0M5JA71 A0A0L0CT42 A0A1W4UJC2 A0A0K8UN29 Q1HRF9 B4LM77 A0A1A9WHW5 Q291B9 B4GAY0 B3ME31 A0A3B0JW41 B4MNS4 A0A182GBC0 B0W577 A0A1Q3FHN9 A0A023EFV0 A0A0L7QQE1 A0A1A9V2M3 A0A1B0C5F3 E0VKS9 A0A084WUK4 A0A182IU65 A0A087TCD1 A0A336MVQ8 T1J8M7 A0A1V9X1Y4 A0A182LT37 A0A1I8Q9C0 A0A182PGD0 A0A1I8M9E0 A0A182YQ94 A0A182RAJ8 A0A182W284 T1DKT4
A0A194Q2G8 A0A2P8ZI11 A0A2J7RQR4 A0A067RD26 A0A0N7Z8Y7 R4G818 A0A0V0G7D0 A0A069DVI1 A0A1B6EFB1 A0A0M8ZVF2 U4UGC3 J3JVP6 A0A1W4WTV8 E2BTE0 R4WQ63 A0A154PEP3 D6WUY3 A0A310SR28 E2ARH1 A0A2A3E0G5 V9IE46 A0A088AK94 A0A0A9Z9B1 A0A1Y1LS78 A0A158NQB1 A0A026WQC8 A0A1B6HSZ2 A0A310SGQ1 A0A151K350 A0A1B6FQ93 K7IQ21 A0A232F8U4 A0A1B6MMI4 E9GS85 A0A195B547 A0A151WGI0 A0A0P5IYV3 A0A0P5PC63 A0A0P4XK11 A0A2R7W450 H9JN51 A0A0T6AX06 A0A0P5FD74 A0A2H8TQD2 A0A2S2PQD1 B4HRN8 A0A0P4WI92 J9JZT8 A0A2S2Q3U3 A0A3Q0JNX5 W8BMX1 A0A1B2AIY1 B3NQT3 B4QFY1 Q8ML92 B4P7N5 B4KNN3 Q16IB6 B4J6L1 A0A0M5JA71 A0A0L0CT42 A0A1W4UJC2 A0A0K8UN29 Q1HRF9 B4LM77 A0A1A9WHW5 Q291B9 B4GAY0 B3ME31 A0A3B0JW41 B4MNS4 A0A182GBC0 B0W577 A0A1Q3FHN9 A0A023EFV0 A0A0L7QQE1 A0A1A9V2M3 A0A1B0C5F3 E0VKS9 A0A084WUK4 A0A182IU65 A0A087TCD1 A0A336MVQ8 T1J8M7 A0A1V9X1Y4 A0A182LT37 A0A1I8Q9C0 A0A182PGD0 A0A1I8M9E0 A0A182YQ94 A0A182RAJ8 A0A182W284 T1DKT4
Pubmed
28756777
23622113
26354079
29403074
24845553
27129103
+ More
26334808 23537049 22516182 20798317 23691247 18362917 19820115 25401762 28004739 21347285 24508170 20075255 28648823 21292972 19121390 17994087 24495485 22936249 10731132 12537572 12537569 16600911 17550304 17510324 26108605 17204158 15632085 26483478 24945155 20566863 24438588 28327890 25315136 25244985
26334808 23537049 22516182 20798317 23691247 18362917 19820115 25401762 28004739 21347285 24508170 20075255 28648823 21292972 19121390 17994087 24495485 22936249 10731132 12537572 12537569 16600911 17550304 17510324 26108605 17204158 15632085 26483478 24945155 20566863 24438588 28327890 25315136 25244985
EMBL
KZ149928
PZC77489.1
ODYU01006654
SOQ48691.1
NWSH01003518
PCG66179.1
+ More
GDQN01004300 JAT86754.1 GAIX01012348 JAA80212.1 KQ461158 KPJ08069.1 KQ459562 KPI99741.1 PYGN01000051 PSN56121.1 NEVH01000618 PNF43180.1 KK852544 KDR21627.1 GDKW01002035 JAI54560.1 ACPB03006195 GAHY01001747 JAA75763.1 GECL01002182 JAP03942.1 GBGD01003570 JAC85319.1 GEDC01000693 JAS36605.1 KQ435827 KOX71930.1 KB632343 ERL93024.1 APGK01058607 BT127314 KB741291 AEE62276.1 ENN70526.1 GL450389 EFN81061.1 AK417831 BAN21046.1 KQ434889 KZC10313.1 KQ971357 EFA08348.1 KQ761453 OAD57543.1 GL442063 EFN63980.1 KZ288487 PBC25230.1 JR039510 AEY58736.1 GBHO01001748 GBHO01001747 GBRD01017225 JAG41856.1 JAG41857.1 JAG48602.1 GEZM01048497 JAV76544.1 ADTU01023014 KK107128 EZA58252.1 GECU01029914 JAS77792.1 KQ761493 OAD57489.1 LKEY01022044 KYN50545.1 GECZ01017402 GECZ01013953 JAS52367.1 JAS55816.1 NNAY01000707 OXU26863.1 GEBQ01002827 JAT37150.1 GL732561 EFX77669.1 KQ976595 KYM79616.1 KQ983176 KYQ46918.1 GDIQ01208621 LRGB01000389 JAK43104.1 KZS19324.1 GDIQ01134898 JAL16828.1 GDIP01243136 JAI80265.1 KK854311 PTY14497.1 BABH01042800 LJIG01022602 KRT79695.1 GDIQ01259872 JAJ91852.1 GFXV01004404 MBW16209.1 GGMR01018986 MBY31605.1 CH480816 EDW47902.1 GDRN01057554 GDRN01057553 JAI65682.1 ABLF02037162 GGMS01003184 MBY72387.1 GAMC01006421 GAMC01006420 GAMC01006418 JAC00138.1 KX531281 ANY27091.1 CH954179 EDV55993.1 CM000362 CM002911 EDX07126.1 KMY93828.1 AE013599 BT001710 CM000158 EDW91061.1 CH933808 EDW10018.1 CH478089 EAT34009.1 CH916367 EDW00914.1 CP012524 ALC41794.1 JRES01000049 KNC34589.1 GDHF01024318 JAI27996.1 DQ440135 ABF18168.1 CH940648 EDW60955.1 CM000071 EAL25193.2 CH479181 EDW32082.1 CH902619 EDV37576.2 OUUW01000001 SPP75288.1 CH963848 EDW73763.1 JXUM01052849 KQ561752 KXJ77634.1 DS231841 EDS34824.1 GFDL01007996 JAV27049.1 GAPW01006124 JAC07474.1 KQ414792 KOC60736.1 JXJN01025955 JXJN01025956 JXJN01025957 DS235250 EEB13985.1 ATLV01027095 KE525423 KFB53898.1 KK114570 KFM62770.1 UFQT01001990 SSX32327.1 JH431957 MNPL01027871 OQR67660.1 AXCM01000968 GAMD01000791 JAB00800.1
GDQN01004300 JAT86754.1 GAIX01012348 JAA80212.1 KQ461158 KPJ08069.1 KQ459562 KPI99741.1 PYGN01000051 PSN56121.1 NEVH01000618 PNF43180.1 KK852544 KDR21627.1 GDKW01002035 JAI54560.1 ACPB03006195 GAHY01001747 JAA75763.1 GECL01002182 JAP03942.1 GBGD01003570 JAC85319.1 GEDC01000693 JAS36605.1 KQ435827 KOX71930.1 KB632343 ERL93024.1 APGK01058607 BT127314 KB741291 AEE62276.1 ENN70526.1 GL450389 EFN81061.1 AK417831 BAN21046.1 KQ434889 KZC10313.1 KQ971357 EFA08348.1 KQ761453 OAD57543.1 GL442063 EFN63980.1 KZ288487 PBC25230.1 JR039510 AEY58736.1 GBHO01001748 GBHO01001747 GBRD01017225 JAG41856.1 JAG41857.1 JAG48602.1 GEZM01048497 JAV76544.1 ADTU01023014 KK107128 EZA58252.1 GECU01029914 JAS77792.1 KQ761493 OAD57489.1 LKEY01022044 KYN50545.1 GECZ01017402 GECZ01013953 JAS52367.1 JAS55816.1 NNAY01000707 OXU26863.1 GEBQ01002827 JAT37150.1 GL732561 EFX77669.1 KQ976595 KYM79616.1 KQ983176 KYQ46918.1 GDIQ01208621 LRGB01000389 JAK43104.1 KZS19324.1 GDIQ01134898 JAL16828.1 GDIP01243136 JAI80265.1 KK854311 PTY14497.1 BABH01042800 LJIG01022602 KRT79695.1 GDIQ01259872 JAJ91852.1 GFXV01004404 MBW16209.1 GGMR01018986 MBY31605.1 CH480816 EDW47902.1 GDRN01057554 GDRN01057553 JAI65682.1 ABLF02037162 GGMS01003184 MBY72387.1 GAMC01006421 GAMC01006420 GAMC01006418 JAC00138.1 KX531281 ANY27091.1 CH954179 EDV55993.1 CM000362 CM002911 EDX07126.1 KMY93828.1 AE013599 BT001710 CM000158 EDW91061.1 CH933808 EDW10018.1 CH478089 EAT34009.1 CH916367 EDW00914.1 CP012524 ALC41794.1 JRES01000049 KNC34589.1 GDHF01024318 JAI27996.1 DQ440135 ABF18168.1 CH940648 EDW60955.1 CM000071 EAL25193.2 CH479181 EDW32082.1 CH902619 EDV37576.2 OUUW01000001 SPP75288.1 CH963848 EDW73763.1 JXUM01052849 KQ561752 KXJ77634.1 DS231841 EDS34824.1 GFDL01007996 JAV27049.1 GAPW01006124 JAC07474.1 KQ414792 KOC60736.1 JXJN01025955 JXJN01025956 JXJN01025957 DS235250 EEB13985.1 ATLV01027095 KE525423 KFB53898.1 KK114570 KFM62770.1 UFQT01001990 SSX32327.1 JH431957 MNPL01027871 OQR67660.1 AXCM01000968 GAMD01000791 JAB00800.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000245037
UP000235965
UP000027135
+ More
UP000015103 UP000053105 UP000030742 UP000019118 UP000192223 UP000008237 UP000076502 UP000007266 UP000000311 UP000242457 UP000005203 UP000005205 UP000053097 UP000078492 UP000002358 UP000215335 UP000000305 UP000078540 UP000075809 UP000076858 UP000005204 UP000001292 UP000007819 UP000079169 UP000008711 UP000000304 UP000000803 UP000002282 UP000009192 UP000008820 UP000001070 UP000092553 UP000037069 UP000192221 UP000008792 UP000091820 UP000001819 UP000008744 UP000007801 UP000268350 UP000007798 UP000069940 UP000249989 UP000002320 UP000053825 UP000078200 UP000092460 UP000009046 UP000030765 UP000075880 UP000054359 UP000192247 UP000075883 UP000095300 UP000075885 UP000095301 UP000076408 UP000075900 UP000075920
UP000015103 UP000053105 UP000030742 UP000019118 UP000192223 UP000008237 UP000076502 UP000007266 UP000000311 UP000242457 UP000005203 UP000005205 UP000053097 UP000078492 UP000002358 UP000215335 UP000000305 UP000078540 UP000075809 UP000076858 UP000005204 UP000001292 UP000007819 UP000079169 UP000008711 UP000000304 UP000000803 UP000002282 UP000009192 UP000008820 UP000001070 UP000092553 UP000037069 UP000192221 UP000008792 UP000091820 UP000001819 UP000008744 UP000007801 UP000268350 UP000007798 UP000069940 UP000249989 UP000002320 UP000053825 UP000078200 UP000092460 UP000009046 UP000030765 UP000075880 UP000054359 UP000192247 UP000075883 UP000095300 UP000075885 UP000095301 UP000076408 UP000075900 UP000075920
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BQX2
A0A2H1W6H4
A0A2A4J2T1
A0A1E1WID3
S4PTB1
A0A0N0PAU7
+ More
A0A194Q2G8 A0A2P8ZI11 A0A2J7RQR4 A0A067RD26 A0A0N7Z8Y7 R4G818 A0A0V0G7D0 A0A069DVI1 A0A1B6EFB1 A0A0M8ZVF2 U4UGC3 J3JVP6 A0A1W4WTV8 E2BTE0 R4WQ63 A0A154PEP3 D6WUY3 A0A310SR28 E2ARH1 A0A2A3E0G5 V9IE46 A0A088AK94 A0A0A9Z9B1 A0A1Y1LS78 A0A158NQB1 A0A026WQC8 A0A1B6HSZ2 A0A310SGQ1 A0A151K350 A0A1B6FQ93 K7IQ21 A0A232F8U4 A0A1B6MMI4 E9GS85 A0A195B547 A0A151WGI0 A0A0P5IYV3 A0A0P5PC63 A0A0P4XK11 A0A2R7W450 H9JN51 A0A0T6AX06 A0A0P5FD74 A0A2H8TQD2 A0A2S2PQD1 B4HRN8 A0A0P4WI92 J9JZT8 A0A2S2Q3U3 A0A3Q0JNX5 W8BMX1 A0A1B2AIY1 B3NQT3 B4QFY1 Q8ML92 B4P7N5 B4KNN3 Q16IB6 B4J6L1 A0A0M5JA71 A0A0L0CT42 A0A1W4UJC2 A0A0K8UN29 Q1HRF9 B4LM77 A0A1A9WHW5 Q291B9 B4GAY0 B3ME31 A0A3B0JW41 B4MNS4 A0A182GBC0 B0W577 A0A1Q3FHN9 A0A023EFV0 A0A0L7QQE1 A0A1A9V2M3 A0A1B0C5F3 E0VKS9 A0A084WUK4 A0A182IU65 A0A087TCD1 A0A336MVQ8 T1J8M7 A0A1V9X1Y4 A0A182LT37 A0A1I8Q9C0 A0A182PGD0 A0A1I8M9E0 A0A182YQ94 A0A182RAJ8 A0A182W284 T1DKT4
A0A194Q2G8 A0A2P8ZI11 A0A2J7RQR4 A0A067RD26 A0A0N7Z8Y7 R4G818 A0A0V0G7D0 A0A069DVI1 A0A1B6EFB1 A0A0M8ZVF2 U4UGC3 J3JVP6 A0A1W4WTV8 E2BTE0 R4WQ63 A0A154PEP3 D6WUY3 A0A310SR28 E2ARH1 A0A2A3E0G5 V9IE46 A0A088AK94 A0A0A9Z9B1 A0A1Y1LS78 A0A158NQB1 A0A026WQC8 A0A1B6HSZ2 A0A310SGQ1 A0A151K350 A0A1B6FQ93 K7IQ21 A0A232F8U4 A0A1B6MMI4 E9GS85 A0A195B547 A0A151WGI0 A0A0P5IYV3 A0A0P5PC63 A0A0P4XK11 A0A2R7W450 H9JN51 A0A0T6AX06 A0A0P5FD74 A0A2H8TQD2 A0A2S2PQD1 B4HRN8 A0A0P4WI92 J9JZT8 A0A2S2Q3U3 A0A3Q0JNX5 W8BMX1 A0A1B2AIY1 B3NQT3 B4QFY1 Q8ML92 B4P7N5 B4KNN3 Q16IB6 B4J6L1 A0A0M5JA71 A0A0L0CT42 A0A1W4UJC2 A0A0K8UN29 Q1HRF9 B4LM77 A0A1A9WHW5 Q291B9 B4GAY0 B3ME31 A0A3B0JW41 B4MNS4 A0A182GBC0 B0W577 A0A1Q3FHN9 A0A023EFV0 A0A0L7QQE1 A0A1A9V2M3 A0A1B0C5F3 E0VKS9 A0A084WUK4 A0A182IU65 A0A087TCD1 A0A336MVQ8 T1J8M7 A0A1V9X1Y4 A0A182LT37 A0A1I8Q9C0 A0A182PGD0 A0A1I8M9E0 A0A182YQ94 A0A182RAJ8 A0A182W284 T1DKT4
PDB
3BS5
E-value=6.20785e-26,
Score=285
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Membrane
Membrane
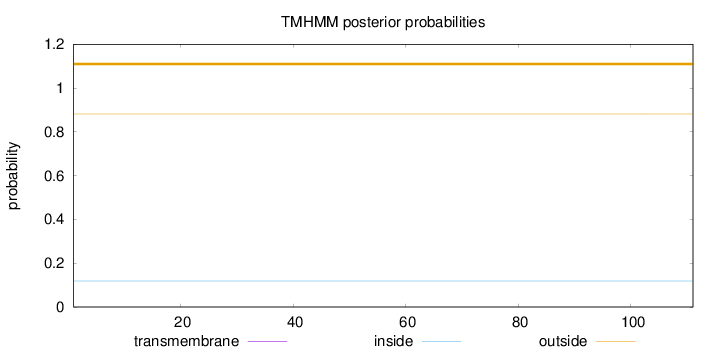
Length:
111
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.11858
outside
1 - 111
Population Genetic Test Statistics
Pi
284.429096
Theta
229.277395
Tajima's D
0.764246
CLR
0.323666
CSRT
0.589970501474926
Interpretation
Uncertain
