Gene
KWMTBOMO01571
Pre Gene Modal
BGIBMGA008838
Annotation
PREDICTED:_protein_msta?_isoform_A-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.884
Sequence
CDS
ATGCTACTTTGTATATTCTGTCAAAAGAAGAGGGGCAAGAGCGATGGACGAAACAAGGAGAATATCTTGCATTCTGACAAAGAAGAAATAGAAGAAGTCTCAACAGCCGATGAGAAGAAAAGCCTATGTTATGAAATCAAATACTCTGATGTTATGGGCAGGTACATTGTGGCAGCAAGAGACATTAAAGCCGGAGAAATCATCATAGAGGAACCGGCTTTAGCTGTTGGACCTTGCACGGGCTGTGCTCTCATATGCCTCGGATGCTACAGGGAAATAGACGAAAATAATATTAACAAATGCAAAAACTGCAAGTGGCCAGTTTGCAGTGGTTCGTGTCCCGGACAAGGAAAATACACAGGTCATTCGACTTACGAATGCGAAACGTTGCGAAGTATACCTCCAGACTACTCTAAGCTTGAAGACTTGAGGGATTCCTACCAAGCATTGATGCCGTTAAGATGTCTACTTTTAAAGAAGGCAGATCCGCCGAAATGGCAGGCTTTATCAGCTATGGAAGCCCACAATGAGCTCCGACGGGCGAGAGGAGATATTTGGCCGACGAACGAGAAGAATGTAGTGCAAAGGATCAAGAGCTGGGGTCTGGAATACGATCCCGAGGAAATACACACTGTCTGTGGGATTTTAGAGGTTAATGCCTTCGAAGTGGGTGGTAGCGGGGCGAACGCTCGTGCTCTTTACGATCGAGCCTATCTCTTGGCCCACGACTGCACTCCCTCCACGACACACACGGACTCCGAGAGGACTCCGGGCAGACCTATAACCGTCAGAGCTTCAATAGCTCATAAAGCCGGAGACTTGATCTCTTTGTGTTACGCTTACACGCTGCAGGGTACACTAAAACGTCGTGAGCACATAAAGCACAGCAAGTTCTTCTGGTGCTCGTGCCGGCGGTGCGCGGACCGCGCCGAGCTCGGCACCTCCGCGTCCGCCTTCCGCTGTCCGCGGTGCGCGGGCGCCGTGCTACCAGAGCGGCCCCTGCAGCGCGACCAGCCCTGGGCCTGCACCCGCGACCACTGCCACTACACCATGCCGGCCGCCGCCGTGCACTTGCTGCTGCAGAGGCTCACAGACGAATACGACCAGATTGACGCGAACGACGTTCCTGGCTTCGAGAATTTCCTCCACAAGTACAGGAACGTGTTGCTGCCCAGTCATTACCTTTGTCTCTCGGCGAAGCACTCCCTCAGTCAGCTGTACGGAAAGGTCACAGACTACATGATACACGAGATGCCCGATACTGAACTGAACAGAAAAATAGATATATGCAGAGATCTGATGAAAGTGTTTGATGTAATCGAACCCGGATATTCGAGGTTGAGAGGTGTAACTCTGTACGAACTACACGCGCCTCTAATGATATTGACAACAAGAGACTTCGAAAAGAAAGCGATAACGAAAGAGAGTCTTCGGAATCGTCTTAAAGAGATAGTACAGTATTTACAAGAATCGGCCCTAATTCTCGGGTTCGAACCAGCGCACTCATCCGAAGGTCTCATGGCGGCGGCCGCAAGAGACGCCCTGAAAAAAATCAAAACATGGGAGCAGATCATCGGCAAGATATCCTGA
Protein
MLLCIFCQKKRGKSDGRNKENILHSDKEEIEEVSTADEKKSLCYEIKYSDVMGRYIVAARDIKAGEIIIEEPALAVGPCTGCALICLGCYREIDENNINKCKNCKWPVCSGSCPGQGKYTGHSTYECETLRSIPPDYSKLEDLRDSYQALMPLRCLLLKKADPPKWQALSAMEAHNELRRARGDIWPTNEKNVVQRIKSWGLEYDPEEIHTVCGILEVNAFEVGGSGANARALYDRAYLLAHDCTPSTTHTDSERTPGRPITVRASIAHKAGDLISLCYAYTLQGTLKRREHIKHSKFFWCSCRRCADRAELGTSASAFRCPRCAGAVLPERPLQRDQPWACTRDHCHYTMPAAAVHLLLQRLTDEYDQIDANDVPGFENFLHKYRNVLLPSHYLCLSAKHSLSQLYGKVTDYMIHEMPDTELNRKIDICRDLMKVFDVIEPGYSRLRGVTLYELHAPLMILTTRDFEKKAITKESLRNRLKEIVQYLQESALILGFEPAHSSEGLMAAAARDALKKIKTWEQIIGKIS
Summary
Uniprot
H9JH41
A0A2A4J8G0
A0A2H1W6H7
A0A2W1BR26
A0A194Q325
A0A0N1PHS6
+ More
A0A212EIW5 A0A1W4WHY5 D6WUY1 U4UGH4 A0A0T6AX24 A0A067RM42 A0A2J7RQQ6 A0A1Y1LWP9 N6UIX5 A0A1B6CES5 A0A336LB95 A0A336N447 A0A336MKR3 A0A1J1HKS4 A0A1L8DX66 A0A1B0D7G8 A0A1B6FLR5 A0A182FUB2 Q17JR4 A0A1S4F084 A0A1J1IRJ1 A0A182G643 A0A023ETM5 A0A1B6KMU9 A0A0L0C386 B0X2T0 A0A1B6DIM2 A0A182M1X4 B4R2Q6 A0A182QX49 A0A182PLA8 Q9VZ41 A0A240PLK3 B3NVC0 A0A1Q3FMT7 A0A1W4UNY3 A0A182WFB9 B4PYV3 A0A182K319 B3MQ91 A0A0R1EBD1 A0A0A1X7X9 E0VN95 B4L719 B4JXB5 A0A1I8MYP4 T1P8T7 A0A182MY56 A0A034V8C4 A0A182XC38 A0A1B0A1B4 A0A0M5JAK7 A0A1A9VWM4 A0A182V4M0 A0A182YNX3 A0A1A9YQE5 A0A1B0AY24 A0A182HND6 A0A0R3NZC6 Q29HZ7 B4MAM9 A0A1I8NZ76 B4H4T1 A0A182LP98 B4MSI4 A0A182GED1 A0A0K8U981 A0A0K8UBP6 A0A1A9WV71 A0A3B0L060 Q7QL17 W8AES0 A0A0K8VM45 W5JJL8 A0A023F6F6 A0A0P6GTM2 A0A0N8B7V8 A0A0N8BMW8 B4IDW9 A0A182TSE0 A0A0N8ARH9 A0A0P5JD04 A0A0N8DXJ7 A0A0P5ZLY7 A0A0N7ZQS5 A0A0P5J7Y8 A0A0P5YS73 A0A0P5PN37 A0A0P5SQ74 A0A0P5PAD1 A0A067R714 A0A0N8EEV6 A0A0P5FIF2 A0A0P6H3U7
A0A212EIW5 A0A1W4WHY5 D6WUY1 U4UGH4 A0A0T6AX24 A0A067RM42 A0A2J7RQQ6 A0A1Y1LWP9 N6UIX5 A0A1B6CES5 A0A336LB95 A0A336N447 A0A336MKR3 A0A1J1HKS4 A0A1L8DX66 A0A1B0D7G8 A0A1B6FLR5 A0A182FUB2 Q17JR4 A0A1S4F084 A0A1J1IRJ1 A0A182G643 A0A023ETM5 A0A1B6KMU9 A0A0L0C386 B0X2T0 A0A1B6DIM2 A0A182M1X4 B4R2Q6 A0A182QX49 A0A182PLA8 Q9VZ41 A0A240PLK3 B3NVC0 A0A1Q3FMT7 A0A1W4UNY3 A0A182WFB9 B4PYV3 A0A182K319 B3MQ91 A0A0R1EBD1 A0A0A1X7X9 E0VN95 B4L719 B4JXB5 A0A1I8MYP4 T1P8T7 A0A182MY56 A0A034V8C4 A0A182XC38 A0A1B0A1B4 A0A0M5JAK7 A0A1A9VWM4 A0A182V4M0 A0A182YNX3 A0A1A9YQE5 A0A1B0AY24 A0A182HND6 A0A0R3NZC6 Q29HZ7 B4MAM9 A0A1I8NZ76 B4H4T1 A0A182LP98 B4MSI4 A0A182GED1 A0A0K8U981 A0A0K8UBP6 A0A1A9WV71 A0A3B0L060 Q7QL17 W8AES0 A0A0K8VM45 W5JJL8 A0A023F6F6 A0A0P6GTM2 A0A0N8B7V8 A0A0N8BMW8 B4IDW9 A0A182TSE0 A0A0N8ARH9 A0A0P5JD04 A0A0N8DXJ7 A0A0P5ZLY7 A0A0N7ZQS5 A0A0P5J7Y8 A0A0P5YS73 A0A0P5PN37 A0A0P5SQ74 A0A0P5PAD1 A0A067R714 A0A0N8EEV6 A0A0P5FIF2 A0A0P6H3U7
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
23537049 24845553 28004739 17510324 26483478 24945155 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25830018 20566863 25315136 25348373 25244985 15632085 20966253 12364791 24495485 20920257 23761445 25474469
23537049 24845553 28004739 17510324 26483478 24945155 26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25830018 20566863 25315136 25348373 25244985 15632085 20966253 12364791 24495485 20920257 23761445 25474469
EMBL
BABH01041036
BABH01041037
NWSH01002406
PCG68365.1
ODYU01006654
SOQ48689.1
+ More
KZ149928 PZC77488.1 KQ459562 KPI99743.1 KQ461158 KPJ08070.1 AGBW02014564 OWR41411.1 KQ971357 EFA08507.1 KB632286 ERL91443.1 LJIG01022602 KRT79705.1 KK852544 KDR21630.1 NEVH01000618 PNF43170.1 GEZM01045051 JAV77952.1 APGK01018201 KB740071 ENN81700.1 GEDC01031647 GEDC01029336 GEDC01025393 GEDC01013625 GEDC01004092 JAS05651.1 JAS07962.1 JAS11905.1 JAS23673.1 JAS33206.1 UFQS01002786 UFQT01002786 SSX14664.1 SSX34061.1 UFQS01004985 UFQT01004985 SSX16646.1 SSX35843.1 UFQT01001113 SSX28967.1 CVRI01000006 CRK88012.1 GFDF01003046 JAV11038.1 AJVK01012426 GECZ01018621 JAS51148.1 CH477231 EAT46910.1 CVRI01000058 CRL02851.1 JXUM01044438 JXUM01044439 JXUM01044440 JXUM01044441 KQ561401 KXJ78702.1 GAPW01000920 JAC12678.1 GEBQ01027217 JAT12760.1 JRES01000955 KNC26808.1 DS232298 EDS39404.1 GEDC01011749 JAS25549.1 AXCM01000948 CM000366 EDX17605.1 AXCN02002034 AE014298 AAF47987.2 CH954180 EDV46108.2 GFDL01006193 JAV28852.1 CM000162 EDX02031.1 CH902621 EDV44517.1 KRK06527.1 GBXI01007276 JAD07016.1 DS235335 EEB14851.1 CH933812 EDW06165.1 CH916376 EDV95391.1 KA645191 AFP59820.1 GAKP01019396 JAC39556.1 CP012528 ALC48974.1 JXJN01005505 APCN01001914 CH379064 KRT06398.1 EAL31609.2 CH940655 EDW66288.2 CH479209 EDW32693.1 CH963851 EDW75073.2 JXUM01057790 KQ561971 KXJ77030.1 GDHF01029166 JAI23148.1 GDHF01028217 GDHF01024193 JAI24097.1 JAI28121.1 OUUW01000028 SPP89768.1 AAAB01008090 EAA03232.3 GAMC01019845 GAMC01019841 JAB86710.1 GDHF01012357 JAI39957.1 ADMH02000957 ETN64572.1 GBBI01001898 JAC16814.1 GDIQ01029719 JAN65018.1 GDIQ01204195 JAK47530.1 GDIQ01162115 GDIQ01160706 JAK89610.1 CH480830 EDW45777.1 GDIQ01250027 JAK01698.1 GDIQ01204242 GDIQ01201858 GDIQ01201857 GDIQ01031065 JAK49868.1 GDIQ01082092 JAN12645.1 GDIP01055387 JAM48328.1 GDIP01221737 JAJ01665.1 GDIQ01201856 GDIQ01094836 JAK49869.1 GDIP01053845 JAM49870.1 GDIQ01147033 JAL04693.1 GDIQ01100999 JAL50727.1 GDIQ01131383 JAL20343.1 KK852699 KDR18208.1 GDIQ01033620 JAN61117.1 GDIQ01254126 JAJ97598.1 GDIQ01035121 JAN59616.1
KZ149928 PZC77488.1 KQ459562 KPI99743.1 KQ461158 KPJ08070.1 AGBW02014564 OWR41411.1 KQ971357 EFA08507.1 KB632286 ERL91443.1 LJIG01022602 KRT79705.1 KK852544 KDR21630.1 NEVH01000618 PNF43170.1 GEZM01045051 JAV77952.1 APGK01018201 KB740071 ENN81700.1 GEDC01031647 GEDC01029336 GEDC01025393 GEDC01013625 GEDC01004092 JAS05651.1 JAS07962.1 JAS11905.1 JAS23673.1 JAS33206.1 UFQS01002786 UFQT01002786 SSX14664.1 SSX34061.1 UFQS01004985 UFQT01004985 SSX16646.1 SSX35843.1 UFQT01001113 SSX28967.1 CVRI01000006 CRK88012.1 GFDF01003046 JAV11038.1 AJVK01012426 GECZ01018621 JAS51148.1 CH477231 EAT46910.1 CVRI01000058 CRL02851.1 JXUM01044438 JXUM01044439 JXUM01044440 JXUM01044441 KQ561401 KXJ78702.1 GAPW01000920 JAC12678.1 GEBQ01027217 JAT12760.1 JRES01000955 KNC26808.1 DS232298 EDS39404.1 GEDC01011749 JAS25549.1 AXCM01000948 CM000366 EDX17605.1 AXCN02002034 AE014298 AAF47987.2 CH954180 EDV46108.2 GFDL01006193 JAV28852.1 CM000162 EDX02031.1 CH902621 EDV44517.1 KRK06527.1 GBXI01007276 JAD07016.1 DS235335 EEB14851.1 CH933812 EDW06165.1 CH916376 EDV95391.1 KA645191 AFP59820.1 GAKP01019396 JAC39556.1 CP012528 ALC48974.1 JXJN01005505 APCN01001914 CH379064 KRT06398.1 EAL31609.2 CH940655 EDW66288.2 CH479209 EDW32693.1 CH963851 EDW75073.2 JXUM01057790 KQ561971 KXJ77030.1 GDHF01029166 JAI23148.1 GDHF01028217 GDHF01024193 JAI24097.1 JAI28121.1 OUUW01000028 SPP89768.1 AAAB01008090 EAA03232.3 GAMC01019845 GAMC01019841 JAB86710.1 GDHF01012357 JAI39957.1 ADMH02000957 ETN64572.1 GBBI01001898 JAC16814.1 GDIQ01029719 JAN65018.1 GDIQ01204195 JAK47530.1 GDIQ01162115 GDIQ01160706 JAK89610.1 CH480830 EDW45777.1 GDIQ01250027 JAK01698.1 GDIQ01204242 GDIQ01201858 GDIQ01201857 GDIQ01031065 JAK49868.1 GDIQ01082092 JAN12645.1 GDIP01055387 JAM48328.1 GDIP01221737 JAJ01665.1 GDIQ01201856 GDIQ01094836 JAK49869.1 GDIP01053845 JAM49870.1 GDIQ01147033 JAL04693.1 GDIQ01100999 JAL50727.1 GDIQ01131383 JAL20343.1 KK852699 KDR18208.1 GDIQ01033620 JAN61117.1 GDIQ01254126 JAJ97598.1 GDIQ01035121 JAN59616.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000192223
+ More
UP000007266 UP000030742 UP000027135 UP000235965 UP000019118 UP000183832 UP000092462 UP000069272 UP000008820 UP000069940 UP000249989 UP000037069 UP000002320 UP000075883 UP000000304 UP000075886 UP000075885 UP000000803 UP000075880 UP000008711 UP000192221 UP000075920 UP000002282 UP000075881 UP000007801 UP000009046 UP000009192 UP000001070 UP000095301 UP000075884 UP000076407 UP000092445 UP000092553 UP000078200 UP000075903 UP000076408 UP000092443 UP000092460 UP000075840 UP000001819 UP000008792 UP000095300 UP000008744 UP000075882 UP000007798 UP000091820 UP000268350 UP000007062 UP000000673 UP000001292 UP000075902
UP000007266 UP000030742 UP000027135 UP000235965 UP000019118 UP000183832 UP000092462 UP000069272 UP000008820 UP000069940 UP000249989 UP000037069 UP000002320 UP000075883 UP000000304 UP000075886 UP000075885 UP000000803 UP000075880 UP000008711 UP000192221 UP000075920 UP000002282 UP000075881 UP000007801 UP000009046 UP000009192 UP000001070 UP000095301 UP000075884 UP000076407 UP000092445 UP000092553 UP000078200 UP000075903 UP000076408 UP000092443 UP000092460 UP000075840 UP000001819 UP000008792 UP000095300 UP000008744 UP000075882 UP000007798 UP000091820 UP000268350 UP000007062 UP000000673 UP000001292 UP000075902
Interpro
SUPFAM
SSF57903
SSF57903
Gene 3D
ProteinModelPortal
H9JH41
A0A2A4J8G0
A0A2H1W6H7
A0A2W1BR26
A0A194Q325
A0A0N1PHS6
+ More
A0A212EIW5 A0A1W4WHY5 D6WUY1 U4UGH4 A0A0T6AX24 A0A067RM42 A0A2J7RQQ6 A0A1Y1LWP9 N6UIX5 A0A1B6CES5 A0A336LB95 A0A336N447 A0A336MKR3 A0A1J1HKS4 A0A1L8DX66 A0A1B0D7G8 A0A1B6FLR5 A0A182FUB2 Q17JR4 A0A1S4F084 A0A1J1IRJ1 A0A182G643 A0A023ETM5 A0A1B6KMU9 A0A0L0C386 B0X2T0 A0A1B6DIM2 A0A182M1X4 B4R2Q6 A0A182QX49 A0A182PLA8 Q9VZ41 A0A240PLK3 B3NVC0 A0A1Q3FMT7 A0A1W4UNY3 A0A182WFB9 B4PYV3 A0A182K319 B3MQ91 A0A0R1EBD1 A0A0A1X7X9 E0VN95 B4L719 B4JXB5 A0A1I8MYP4 T1P8T7 A0A182MY56 A0A034V8C4 A0A182XC38 A0A1B0A1B4 A0A0M5JAK7 A0A1A9VWM4 A0A182V4M0 A0A182YNX3 A0A1A9YQE5 A0A1B0AY24 A0A182HND6 A0A0R3NZC6 Q29HZ7 B4MAM9 A0A1I8NZ76 B4H4T1 A0A182LP98 B4MSI4 A0A182GED1 A0A0K8U981 A0A0K8UBP6 A0A1A9WV71 A0A3B0L060 Q7QL17 W8AES0 A0A0K8VM45 W5JJL8 A0A023F6F6 A0A0P6GTM2 A0A0N8B7V8 A0A0N8BMW8 B4IDW9 A0A182TSE0 A0A0N8ARH9 A0A0P5JD04 A0A0N8DXJ7 A0A0P5ZLY7 A0A0N7ZQS5 A0A0P5J7Y8 A0A0P5YS73 A0A0P5PN37 A0A0P5SQ74 A0A0P5PAD1 A0A067R714 A0A0N8EEV6 A0A0P5FIF2 A0A0P6H3U7
A0A212EIW5 A0A1W4WHY5 D6WUY1 U4UGH4 A0A0T6AX24 A0A067RM42 A0A2J7RQQ6 A0A1Y1LWP9 N6UIX5 A0A1B6CES5 A0A336LB95 A0A336N447 A0A336MKR3 A0A1J1HKS4 A0A1L8DX66 A0A1B0D7G8 A0A1B6FLR5 A0A182FUB2 Q17JR4 A0A1S4F084 A0A1J1IRJ1 A0A182G643 A0A023ETM5 A0A1B6KMU9 A0A0L0C386 B0X2T0 A0A1B6DIM2 A0A182M1X4 B4R2Q6 A0A182QX49 A0A182PLA8 Q9VZ41 A0A240PLK3 B3NVC0 A0A1Q3FMT7 A0A1W4UNY3 A0A182WFB9 B4PYV3 A0A182K319 B3MQ91 A0A0R1EBD1 A0A0A1X7X9 E0VN95 B4L719 B4JXB5 A0A1I8MYP4 T1P8T7 A0A182MY56 A0A034V8C4 A0A182XC38 A0A1B0A1B4 A0A0M5JAK7 A0A1A9VWM4 A0A182V4M0 A0A182YNX3 A0A1A9YQE5 A0A1B0AY24 A0A182HND6 A0A0R3NZC6 Q29HZ7 B4MAM9 A0A1I8NZ76 B4H4T1 A0A182LP98 B4MSI4 A0A182GED1 A0A0K8U981 A0A0K8UBP6 A0A1A9WV71 A0A3B0L060 Q7QL17 W8AES0 A0A0K8VM45 W5JJL8 A0A023F6F6 A0A0P6GTM2 A0A0N8B7V8 A0A0N8BMW8 B4IDW9 A0A182TSE0 A0A0N8ARH9 A0A0P5JD04 A0A0N8DXJ7 A0A0P5ZLY7 A0A0N7ZQS5 A0A0P5J7Y8 A0A0P5YS73 A0A0P5PN37 A0A0P5SQ74 A0A0P5PAD1 A0A067R714 A0A0N8EEV6 A0A0P5FIF2 A0A0P6H3U7
PDB
6FND
E-value=0.0109519,
Score=92
Ontologies
GO
Topology
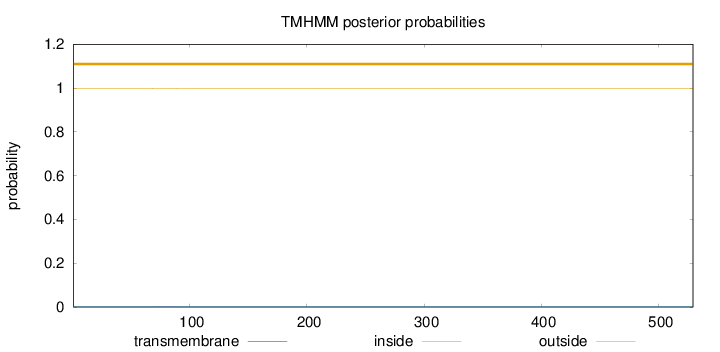
Length:
529
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00774999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00173
outside
1 - 529
Population Genetic Test Statistics
Pi
161.742703
Theta
143.488171
Tajima's D
0.561743
CLR
0.05762
CSRT
0.535323233838308
Interpretation
Uncertain
