Gene
KWMTBOMO01570
Pre Gene Modal
BGIBMGA008839
Annotation
PREDICTED:_protein_msta?_isoform_B_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.333 Nuclear Reliability : 1.777
Sequence
CDS
ATGGTAGAACAGGGTTCTTGTGCAGTTTGTCTATCGCCTGCGCCCCAAAAATGTTCAGGATGTCAGTACGTCTACTACTGTTCTAAAGAACATCAGAAACAAGATTGGAAGCAGCACAAATACCAGTGCTCCCCAGTACGAGTGAAAGAAGACTCGCAATTCGGGAGGTTTCTCGAGGCCACGCGAGAAATAAAATCTGGTGATATTATCTTGAAGGAGAAACCATTGATATCGGGACCTGCGCAGGTGACTCCACCAGTATGTCTCGGCTGCTACAAATTGCTGGAAGAAGGAAAGACTGTGAAATGTAGCAAGTGTGGTTGGCCGTTCTGTTCTGAGCTGTGCACTGAGAAACCAGAACACGAACCTGAATGTCATTATACGGAAAAAAGAGGAGAAAAGGTCAACATAACAACATTTGGTTCGCCACATCCGAACTATCAATGCATCACCGTCCTCCGATGCTTGTACCAGCGGGACCACAATGAGCCGTTGTGGAAGAAGCTCCAAGCGTTACAGTCTCACGACGAGGACAGGAGGGGCACAGACAAGTGGAATAACGATAAAAAGATGATCTGCGATTTCATATGGATGTTCTTCAAACTTGACGGCGTCTTCAACGAGGAAGAAATAATGAAATGCTGTGGGATTTTGCAGATCAACGGGCATGAAGTACCTTTAATGGAGCCGGAGTATCTGGCAGTATACGACCGCATCTCCATGGTCGAACACAACTGTCGCGCCAACTGTAACAAAAGTTTTACGTCTGCCGGAGAAATTATCCTGTGTGCTGGAGACAGTATCAAGACTGGCGAGCATGTAACAGTCTGCTACACAGACCCCTTGTGGGGCACGGAAGCGCGGCGCCACCACCTTGCCGACTCCAAGTTCTTCGAATGCAACTGTGAGAGATGCTCTGACGTCACCGAACTCGCGACAATGTACAGTGCTGTCAAATGTAAAAAGAAGGACTGCAAAGGCTATCTTCTACCGGAAACTTTCATCGTACCGCTCTTGCACAAGACGAAGAAACCTGATCCTGAAACTCGTAACTTAGACAGAAAGTTCTGGATTTGTAATAGCTGCAAGGACGCCGTATCGGATGCCATTATACAACAACTCCTGCAGGATATTGGACGAGAACTGAGCGTTATGCCAAAGGGTGAAGCTGATGCTTGCGAAAGGTAA
Protein
MVEQGSCAVCLSPAPQKCSGCQYVYYCSKEHQKQDWKQHKYQCSPVRVKEDSQFGRFLEATREIKSGDIILKEKPLISGPAQVTPPVCLGCYKLLEEGKTVKCSKCGWPFCSELCTEKPEHEPECHYTEKRGEKVNITTFGSPHPNYQCITVLRCLYQRDHNEPLWKKLQALQSHDEDRRGTDKWNNDKKMICDFIWMFFKLDGVFNEEEIMKCCGILQINGHEVPLMEPEYLAVYDRISMVEHNCRANCNKSFTSAGEIILCAGDSIKTGEHVTVCYTDPLWGTEARRHHLADSKFFECNCERCSDVTELATMYSAVKCKKKDCKGYLLPETFIVPLLHKTKKPDPETRNLDRKFWICNSCKDAVSDAIIQQLLQDIGRELSVMPKGEADACER
Summary
Uniprot
H9JH42
A0A2A4JA02
A0A2W1BWA6
A0A2H1W6K0
A0A194Q405
A0A0N1INM4
+ More
A0A212FA54 A0A067RCS6 A0A1W4WG27 A0A139WDX7 T1I1Y7 V5I9L0 U4TWJ2 N6UBC5 A0A1B6KW78 U4TWD0 N6TQN3 A0A084VRL9 A0A336MFT2 A0A182HND0 Q7QGG8 A0A182LP89 A0A182VMT4 A0A336MSP7 A0A336MII8 A0A182XC32 A0A1B6LJ04 A0A182U585 A0A1B6LFN0 A0A1B6JIQ3 A0A182QTK0 A0A182Y5X0 A0A182FUB9 A0A182JVG9 A0A182RQZ4 A0A182J9R3 A0A182W2G3 A0A1Q3G585 A0A182MD85 A0A182MY62 A0A182GEC9 A0A023EZV5 A0A1S4F0F0 Q17JR2 A0A154PC23 A0A146KWH5 A0A3Q0IYE9 A0A1A9WNC5 A0A1Y1N7B3 A0A088A9C0 A0A0L0CBP4 A0A1Y1N7Z3 A0A1B0AKC5 A0A1A9V7P1 A0A0A9Y6I6 A0A232EFX9 K7J0F2 A0A1I8MXW6 W8C7K8 A0A034WPA2 A0A3B0JMW8 A0A182PU93 B4H526 B4P730 B4JA05 A0A1A9XPS2 B4KRB6 B5E1F5 B3NR70 A0A2A3EFI2 A0A0K8WET2 A0A1B0AM53 A0A0M4EFV0 Q7K561 A0A0J9U1R8 B4HRA0 A0A1W4WA57 A0A0A1XD82 B3MDY3 B4MJN1 A0A2S2Q9E4 A0A1B0D7G9 A0A2P8ZHZ6 B4LLD4 A0A2H8TU54 W5JCL7 A0A0T6B9M4 A0A1J1I0J5 J9JY07 A0A2S2PQ37 A0A1B0GHX9 W5JT41 A0A0K2TV79 A0A2S2N781 A0A1B6CC67 A0A0L7QM18 A0A182FLM7 A0A2P2I598 U5EVB9 E9G7C4
A0A212FA54 A0A067RCS6 A0A1W4WG27 A0A139WDX7 T1I1Y7 V5I9L0 U4TWJ2 N6UBC5 A0A1B6KW78 U4TWD0 N6TQN3 A0A084VRL9 A0A336MFT2 A0A182HND0 Q7QGG8 A0A182LP89 A0A182VMT4 A0A336MSP7 A0A336MII8 A0A182XC32 A0A1B6LJ04 A0A182U585 A0A1B6LFN0 A0A1B6JIQ3 A0A182QTK0 A0A182Y5X0 A0A182FUB9 A0A182JVG9 A0A182RQZ4 A0A182J9R3 A0A182W2G3 A0A1Q3G585 A0A182MD85 A0A182MY62 A0A182GEC9 A0A023EZV5 A0A1S4F0F0 Q17JR2 A0A154PC23 A0A146KWH5 A0A3Q0IYE9 A0A1A9WNC5 A0A1Y1N7B3 A0A088A9C0 A0A0L0CBP4 A0A1Y1N7Z3 A0A1B0AKC5 A0A1A9V7P1 A0A0A9Y6I6 A0A232EFX9 K7J0F2 A0A1I8MXW6 W8C7K8 A0A034WPA2 A0A3B0JMW8 A0A182PU93 B4H526 B4P730 B4JA05 A0A1A9XPS2 B4KRB6 B5E1F5 B3NR70 A0A2A3EFI2 A0A0K8WET2 A0A1B0AM53 A0A0M4EFV0 Q7K561 A0A0J9U1R8 B4HRA0 A0A1W4WA57 A0A0A1XD82 B3MDY3 B4MJN1 A0A2S2Q9E4 A0A1B0D7G9 A0A2P8ZHZ6 B4LLD4 A0A2H8TU54 W5JCL7 A0A0T6B9M4 A0A1J1I0J5 J9JY07 A0A2S2PQ37 A0A1B0GHX9 W5JT41 A0A0K2TV79 A0A2S2N781 A0A1B6CC67 A0A0L7QM18 A0A182FLM7 A0A2P2I598 U5EVB9 E9G7C4
Pubmed
19121390
28756777
26354079
22118469
24845553
18362917
+ More
19820115 23537049 24438588 12364791 20966253 25244985 26483478 25474469 17510324 26823975 28004739 26108605 25401762 28648823 20075255 25315136 24495485 25348373 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 29403074 20920257 23761445 21292972
19820115 23537049 24438588 12364791 20966253 25244985 26483478 25474469 17510324 26823975 28004739 26108605 25401762 28648823 20075255 25315136 24495485 25348373 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 29403074 20920257 23761445 21292972
EMBL
BABH01041038
BABH01041039
BABH01041040
NWSH01002406
PCG68364.1
KZ149928
+ More
PZC77487.1 ODYU01006654 SOQ48688.1 KQ459562 KPI99744.1 KQ461158 KPJ08071.1 AGBW02009521 OWR50603.1 KK852544 KDR21631.1 KQ971357 KYB26114.1 ACPB03006195 GALX01002750 JAB65716.1 KB631763 ERL85939.1 APGK01035775 APGK01035776 APGK01035777 APGK01035778 APGK01035779 APGK01035780 KB740928 ENN77956.1 GEBQ01024270 JAT15707.1 KB631759 ERL85859.1 APGK01058762 KB741291 ENN70601.1 ATLV01015746 KE525033 KFB40613.1 UFQT01001171 SSX29262.1 APCN01001905 AAAB01008834 EAA05706.4 UFQT01001115 SSX28968.1 SSX29261.1 GEBQ01016337 JAT23640.1 GEBQ01017492 JAT22485.1 GECU01008649 JAS99057.1 AXCN02002036 GFDL01000075 JAV34970.1 AXCM01010579 JXUM01057778 JXUM01057779 KQ561971 KXJ77028.1 GBBI01003920 JAC14792.1 CH477231 EAT46912.1 KQ434869 KZC09337.1 GDHC01019489 JAP99139.1 GEZM01011203 JAV93599.1 JRES01000644 KNC29660.1 GEZM01011204 JAV93598.1 GBHO01018494 JAG25110.1 NNAY01004901 OXU17269.1 GAMC01007951 JAB98604.1 GAKP01002478 JAC56474.1 OUUW01000001 SPP74666.1 CH479210 EDW32862.1 CM000158 EDW90995.1 CH916367 EDW02592.1 CH933808 EDW09332.1 CM000071 EDY69496.2 CH954179 EDV56059.1 KZ288259 PBC30497.1 GDHF01002707 JAI49607.1 JXJN01000335 CP012524 ALC41160.1 AE013599 AY051441 AAF58272.1 AAK92865.1 CM002911 KMY93730.1 CH480816 EDW47829.1 GBXI01005391 JAD08901.1 CH902619 EDV37528.1 CH963846 EDW72320.1 GGMS01005141 MBY74344.1 AJVK01012430 AJVK01012431 AJVK01012432 PYGN01000051 PSN56125.1 CH940648 EDW61886.1 GFXV01005979 MBW17784.1 ADMH02001793 ETN61078.1 LJIG01002898 KRT84042.1 CVRI01000036 CRK93100.1 ABLF02010518 GGMR01018963 MBY31582.1 AJWK01010431 ADMH02000225 ETN67311.1 HACA01011935 CDW29296.1 GGMR01000370 MBY12989.1 GEDC01026241 GEDC01017743 JAS11057.1 JAS19555.1 KQ414902 KOC59662.1 IACF01003444 LAB69060.1 GANO01003474 JAB56397.1 GL732534 EFX84442.1
PZC77487.1 ODYU01006654 SOQ48688.1 KQ459562 KPI99744.1 KQ461158 KPJ08071.1 AGBW02009521 OWR50603.1 KK852544 KDR21631.1 KQ971357 KYB26114.1 ACPB03006195 GALX01002750 JAB65716.1 KB631763 ERL85939.1 APGK01035775 APGK01035776 APGK01035777 APGK01035778 APGK01035779 APGK01035780 KB740928 ENN77956.1 GEBQ01024270 JAT15707.1 KB631759 ERL85859.1 APGK01058762 KB741291 ENN70601.1 ATLV01015746 KE525033 KFB40613.1 UFQT01001171 SSX29262.1 APCN01001905 AAAB01008834 EAA05706.4 UFQT01001115 SSX28968.1 SSX29261.1 GEBQ01016337 JAT23640.1 GEBQ01017492 JAT22485.1 GECU01008649 JAS99057.1 AXCN02002036 GFDL01000075 JAV34970.1 AXCM01010579 JXUM01057778 JXUM01057779 KQ561971 KXJ77028.1 GBBI01003920 JAC14792.1 CH477231 EAT46912.1 KQ434869 KZC09337.1 GDHC01019489 JAP99139.1 GEZM01011203 JAV93599.1 JRES01000644 KNC29660.1 GEZM01011204 JAV93598.1 GBHO01018494 JAG25110.1 NNAY01004901 OXU17269.1 GAMC01007951 JAB98604.1 GAKP01002478 JAC56474.1 OUUW01000001 SPP74666.1 CH479210 EDW32862.1 CM000158 EDW90995.1 CH916367 EDW02592.1 CH933808 EDW09332.1 CM000071 EDY69496.2 CH954179 EDV56059.1 KZ288259 PBC30497.1 GDHF01002707 JAI49607.1 JXJN01000335 CP012524 ALC41160.1 AE013599 AY051441 AAF58272.1 AAK92865.1 CM002911 KMY93730.1 CH480816 EDW47829.1 GBXI01005391 JAD08901.1 CH902619 EDV37528.1 CH963846 EDW72320.1 GGMS01005141 MBY74344.1 AJVK01012430 AJVK01012431 AJVK01012432 PYGN01000051 PSN56125.1 CH940648 EDW61886.1 GFXV01005979 MBW17784.1 ADMH02001793 ETN61078.1 LJIG01002898 KRT84042.1 CVRI01000036 CRK93100.1 ABLF02010518 GGMR01018963 MBY31582.1 AJWK01010431 ADMH02000225 ETN67311.1 HACA01011935 CDW29296.1 GGMR01000370 MBY12989.1 GEDC01026241 GEDC01017743 JAS11057.1 JAS19555.1 KQ414902 KOC59662.1 IACF01003444 LAB69060.1 GANO01003474 JAB56397.1 GL732534 EFX84442.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000192223 UP000007266 UP000015103 UP000030742 UP000019118 UP000030765 UP000075840 UP000007062 UP000075882 UP000075903 UP000076407 UP000075902 UP000075886 UP000076408 UP000069272 UP000075881 UP000075900 UP000075880 UP000075920 UP000075883 UP000075884 UP000069940 UP000249989 UP000008820 UP000076502 UP000079169 UP000091820 UP000005203 UP000037069 UP000092445 UP000078200 UP000215335 UP000002358 UP000095301 UP000268350 UP000075885 UP000008744 UP000002282 UP000001070 UP000092443 UP000009192 UP000001819 UP000008711 UP000242457 UP000092460 UP000092553 UP000000803 UP000001292 UP000192221 UP000007801 UP000007798 UP000092462 UP000245037 UP000008792 UP000000673 UP000183832 UP000007819 UP000092461 UP000053825 UP000000305
UP000192223 UP000007266 UP000015103 UP000030742 UP000019118 UP000030765 UP000075840 UP000007062 UP000075882 UP000075903 UP000076407 UP000075902 UP000075886 UP000076408 UP000069272 UP000075881 UP000075900 UP000075880 UP000075920 UP000075883 UP000075884 UP000069940 UP000249989 UP000008820 UP000076502 UP000079169 UP000091820 UP000005203 UP000037069 UP000092445 UP000078200 UP000215335 UP000002358 UP000095301 UP000268350 UP000075885 UP000008744 UP000002282 UP000001070 UP000092443 UP000009192 UP000001819 UP000008711 UP000242457 UP000092460 UP000092553 UP000000803 UP000001292 UP000192221 UP000007801 UP000007798 UP000092462 UP000245037 UP000008792 UP000000673 UP000183832 UP000007819 UP000092461 UP000053825 UP000000305
Interpro
Gene 3D
ProteinModelPortal
H9JH42
A0A2A4JA02
A0A2W1BWA6
A0A2H1W6K0
A0A194Q405
A0A0N1INM4
+ More
A0A212FA54 A0A067RCS6 A0A1W4WG27 A0A139WDX7 T1I1Y7 V5I9L0 U4TWJ2 N6UBC5 A0A1B6KW78 U4TWD0 N6TQN3 A0A084VRL9 A0A336MFT2 A0A182HND0 Q7QGG8 A0A182LP89 A0A182VMT4 A0A336MSP7 A0A336MII8 A0A182XC32 A0A1B6LJ04 A0A182U585 A0A1B6LFN0 A0A1B6JIQ3 A0A182QTK0 A0A182Y5X0 A0A182FUB9 A0A182JVG9 A0A182RQZ4 A0A182J9R3 A0A182W2G3 A0A1Q3G585 A0A182MD85 A0A182MY62 A0A182GEC9 A0A023EZV5 A0A1S4F0F0 Q17JR2 A0A154PC23 A0A146KWH5 A0A3Q0IYE9 A0A1A9WNC5 A0A1Y1N7B3 A0A088A9C0 A0A0L0CBP4 A0A1Y1N7Z3 A0A1B0AKC5 A0A1A9V7P1 A0A0A9Y6I6 A0A232EFX9 K7J0F2 A0A1I8MXW6 W8C7K8 A0A034WPA2 A0A3B0JMW8 A0A182PU93 B4H526 B4P730 B4JA05 A0A1A9XPS2 B4KRB6 B5E1F5 B3NR70 A0A2A3EFI2 A0A0K8WET2 A0A1B0AM53 A0A0M4EFV0 Q7K561 A0A0J9U1R8 B4HRA0 A0A1W4WA57 A0A0A1XD82 B3MDY3 B4MJN1 A0A2S2Q9E4 A0A1B0D7G9 A0A2P8ZHZ6 B4LLD4 A0A2H8TU54 W5JCL7 A0A0T6B9M4 A0A1J1I0J5 J9JY07 A0A2S2PQ37 A0A1B0GHX9 W5JT41 A0A0K2TV79 A0A2S2N781 A0A1B6CC67 A0A0L7QM18 A0A182FLM7 A0A2P2I598 U5EVB9 E9G7C4
A0A212FA54 A0A067RCS6 A0A1W4WG27 A0A139WDX7 T1I1Y7 V5I9L0 U4TWJ2 N6UBC5 A0A1B6KW78 U4TWD0 N6TQN3 A0A084VRL9 A0A336MFT2 A0A182HND0 Q7QGG8 A0A182LP89 A0A182VMT4 A0A336MSP7 A0A336MII8 A0A182XC32 A0A1B6LJ04 A0A182U585 A0A1B6LFN0 A0A1B6JIQ3 A0A182QTK0 A0A182Y5X0 A0A182FUB9 A0A182JVG9 A0A182RQZ4 A0A182J9R3 A0A182W2G3 A0A1Q3G585 A0A182MD85 A0A182MY62 A0A182GEC9 A0A023EZV5 A0A1S4F0F0 Q17JR2 A0A154PC23 A0A146KWH5 A0A3Q0IYE9 A0A1A9WNC5 A0A1Y1N7B3 A0A088A9C0 A0A0L0CBP4 A0A1Y1N7Z3 A0A1B0AKC5 A0A1A9V7P1 A0A0A9Y6I6 A0A232EFX9 K7J0F2 A0A1I8MXW6 W8C7K8 A0A034WPA2 A0A3B0JMW8 A0A182PU93 B4H526 B4P730 B4JA05 A0A1A9XPS2 B4KRB6 B5E1F5 B3NR70 A0A2A3EFI2 A0A0K8WET2 A0A1B0AM53 A0A0M4EFV0 Q7K561 A0A0J9U1R8 B4HRA0 A0A1W4WA57 A0A0A1XD82 B3MDY3 B4MJN1 A0A2S2Q9E4 A0A1B0D7G9 A0A2P8ZHZ6 B4LLD4 A0A2H8TU54 W5JCL7 A0A0T6B9M4 A0A1J1I0J5 J9JY07 A0A2S2PQ37 A0A1B0GHX9 W5JT41 A0A0K2TV79 A0A2S2N781 A0A1B6CC67 A0A0L7QM18 A0A182FLM7 A0A2P2I598 U5EVB9 E9G7C4
PDB
5HQ8
E-value=3.64066e-10,
Score=156
Ontologies
GO
PANTHER
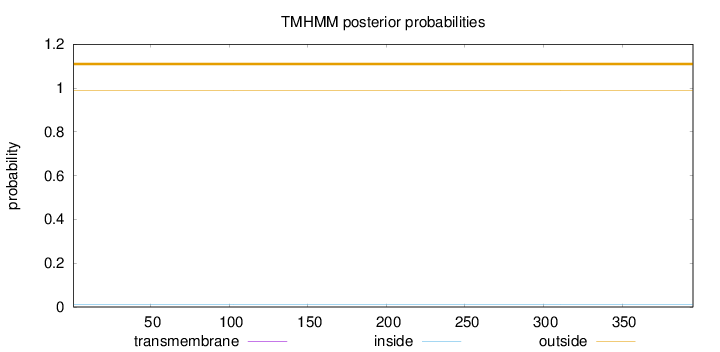
Topology
Length:
395
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.01088
outside
1 - 395
Population Genetic Test Statistics
Pi
156.72939
Theta
153.665328
Tajima's D
0.083986
CLR
100.964578
CSRT
0.397730113494325
Interpretation
Possibly Positive selection
