Gene
KWMTBOMO01555
Pre Gene Modal
BGIBMGA009078
Annotation
PREDICTED:_potassium_voltage-gated_channel_protein_Shaw-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.251
Sequence
CDS
ATGGATGTCGTGGTAGAGCTAACAATAGTTAGACAGTACGTTGATAAAGCAGAGATTTTGGAGTTTATCTCTATCATCAAGATCCTGAGGCTGTTCAAGCTGACCAGACACAGTCCAGGGCTGAAGATCTTGGTGCACACCTTCAAGGCGTCAGCAAAGGAACTGACGCTGTTAGTGTTTTTTCTGGTCCTCGGAATCGTGATCTTCGCGAGCTTGGTGTACTACGCTGAGAAATCGCAGGATAACCCAGAGAACCAGTTTAAATCTATACCCCTCGGTCTTTGGTGGGCGATCGTCACGATGACCACTGTCGGGTATGGAGATATGGCGCCCAAGACTTACCTCGGGATGTTTGTGGGCGCTCTCTGCGCCTTGGCTGGAGTGCTCACTATCGCGCTGCCCGTTCCCGTGATAGTATCCAACTTCTCCATGTTCTACTCCCATACTCAGGCAAGTGGATAG
Protein
MDVVVELTIVRQYVDKAEILEFISIIKILRLFKLTRHSPGLKILVHTFKASAKELTLLVFFLVLGIVIFASLVYYAEKSQDNPENQFKSIPLGLWWAIVTMTTVGYGDMAPKTYLGMFVGALCALAGVLTIALPVPVIVSNFSMFYSHTQASG
Summary
Similarity
Belongs to the potassium channel family.
Uniprot
A0A2W1C080
A0A194Q339
A0A0N0PCM8
A0A2S2NNR1
A0A182KDV1
A0A2S2Q878
+ More
A0A0A9Y0S3 A0A0A9Y3U7 A0A0T6BEL8 A0A0P5GGB7 A0A0P5KSL0 A0A0P5FU90 A0A0P4Y7U0 A0A182JLR5 A0A0P5RN71 A0A0P5BJ70 A0A0P5G9Y4 E9G6G5 A0A182L169 A0A0P6ELD9 A0A0P6CNE0 A0A0P6FHY7 A0A0P5VYS4 A0A0P5PSS9 A0A2S2Q771 A0A2J7RQK8 A0A0P5BSD2 A0A1B0D0Z9 A0A0P5MTL3 A0A0P5WJQ2 A0A2H8TU10 J9KBA4 A0A0P5CVD5 A0A0P5YDY4 A0A0P6FFB6 A0A1B6JVD2 A0A067RPK6 A0A0P5PD88 A0A0P4YND9 A0A0P4Z2A9 A0A0P5L222 A0A0P6JTA4 A0A0P6G807 E9G2I2 A0A182PPR6 A0A0P4Z728 A0A182HUF7 A0A182XKZ8 A0A182SFK4 A0A0P5ARG3 A0A0N7ZJ77 E9J3X3 Q7PJ62 A0A182L172 A0A0P4X5J6 A0A0P5M6G9 A0A0P5MM19 A0A0P5PAC5 T1G8N9 A0A182R4S3 A0A1Y1MBB0 A0A0P5ABL6 U4TY14 A0A0P5H9G8 A0A0P6IH24 A0A084VN64 A0A023GDU2 A0A1S4GXL1 A0A0N8CYR5 A0A182Y3M5 A0A0P5TWV6 A0A0N8AFK0 A0A0P5N395 A0A336KTB5 A0A182F5V3 A0A1Y3AWI0 A0A0P6CAE6 A0A0P6GJM9 A0A0P5YJA6 A0A0N8CCA5 N6U4K5 A0A0P5ZEA9 A0A0P5Z2I1 A0A0P5XHT2 A0A2Y9D1K8 A0A0P6IC74 A0A023FQ08 A0A182M9X6 A0A2P8ZHZ7 A0A087T9Q1 A0A2T7PGE8 A0A182F5U9 A0A182K7C2 A0A0P5R5I5 A0A182PPR2 T1KJ18 A0A1E1X451 A0A2A4JJ74 A0A182XKZ3 A0A2A4JKU0
A0A0A9Y0S3 A0A0A9Y3U7 A0A0T6BEL8 A0A0P5GGB7 A0A0P5KSL0 A0A0P5FU90 A0A0P4Y7U0 A0A182JLR5 A0A0P5RN71 A0A0P5BJ70 A0A0P5G9Y4 E9G6G5 A0A182L169 A0A0P6ELD9 A0A0P6CNE0 A0A0P6FHY7 A0A0P5VYS4 A0A0P5PSS9 A0A2S2Q771 A0A2J7RQK8 A0A0P5BSD2 A0A1B0D0Z9 A0A0P5MTL3 A0A0P5WJQ2 A0A2H8TU10 J9KBA4 A0A0P5CVD5 A0A0P5YDY4 A0A0P6FFB6 A0A1B6JVD2 A0A067RPK6 A0A0P5PD88 A0A0P4YND9 A0A0P4Z2A9 A0A0P5L222 A0A0P6JTA4 A0A0P6G807 E9G2I2 A0A182PPR6 A0A0P4Z728 A0A182HUF7 A0A182XKZ8 A0A182SFK4 A0A0P5ARG3 A0A0N7ZJ77 E9J3X3 Q7PJ62 A0A182L172 A0A0P4X5J6 A0A0P5M6G9 A0A0P5MM19 A0A0P5PAC5 T1G8N9 A0A182R4S3 A0A1Y1MBB0 A0A0P5ABL6 U4TY14 A0A0P5H9G8 A0A0P6IH24 A0A084VN64 A0A023GDU2 A0A1S4GXL1 A0A0N8CYR5 A0A182Y3M5 A0A0P5TWV6 A0A0N8AFK0 A0A0P5N395 A0A336KTB5 A0A182F5V3 A0A1Y3AWI0 A0A0P6CAE6 A0A0P6GJM9 A0A0P5YJA6 A0A0N8CCA5 N6U4K5 A0A0P5ZEA9 A0A0P5Z2I1 A0A0P5XHT2 A0A2Y9D1K8 A0A0P6IC74 A0A023FQ08 A0A182M9X6 A0A2P8ZHZ7 A0A087T9Q1 A0A2T7PGE8 A0A182F5U9 A0A182K7C2 A0A0P5R5I5 A0A182PPR2 T1KJ18 A0A1E1X451 A0A2A4JJ74 A0A182XKZ3 A0A2A4JKU0
Pubmed
EMBL
KZ149928
PZC77473.1
KQ459562
KPI99758.1
KQ460636
KPJ13360.1
+ More
GGMR01006175 MBY18794.1 GGMS01004721 MBY73924.1 GBHO01017815 JAG25789.1 GBHO01017816 JAG25788.1 LJIG01001161 KRT85779.1 GDIQ01248423 JAK03302.1 GDIQ01205639 JAK46086.1 GDIQ01249975 JAK01750.1 GDIP01232326 JAI91075.1 GDIQ01107418 GDIP01059485 JAL44308.1 JAM44230.1 GDIP01183816 JAJ39586.1 GDIQ01251468 JAK00257.1 GL732533 EFX84931.1 GDIQ01068031 JAN26706.1 GDIQ01089024 JAN05713.1 GDIQ01049952 JAN44785.1 GDIP01094347 JAM09368.1 GDIQ01144903 JAL06823.1 GGMS01004371 MBY73574.1 NEVH01001335 PNF43116.1 GDIP01185110 JAJ38292.1 AJVK01021811 GDIQ01174096 JAK77629.1 GDIQ01112920 GDIP01085480 LRGB01002384 JAL38806.1 JAM18235.1 KZS07935.1 GFXV01005625 MBW17430.1 ABLF02040673 ABLF02040674 ABLF02040675 ABLF02040682 ABLF02040683 ABLF02040686 ABLF02040688 ABLF02040693 GDIP01165885 JAJ57517.1 GDIP01073234 JAM30481.1 GDIQ01048466 JAN46271.1 GECU01004560 JAT03147.1 KK852544 KDR21624.1 GDIQ01139812 JAL11914.1 GDIP01226307 JAI97094.1 GDIP01220166 JAJ03236.1 GDIQ01176393 JAK75332.1 GDIQ01009264 JAN85473.1 GDIQ01048465 JAN46272.1 GL732530 EFX86305.1 GDIP01218581 JAJ04821.1 APCN01002460 GDIP01199931 JAJ23471.1 GDIP01240121 JAI83280.1 GL768069 EFZ12524.1 AAAB01008964 EAA43883.4 GDIP01247494 JAI75907.1 GDIQ01160111 JAK91614.1 GDIQ01161666 JAK90059.1 GDIQ01137465 JAL14261.1 AMQM01010917 KB095994 ESO09179.1 GEZM01035706 GEZM01035705 JAV83119.1 GDIP01204709 JAJ18693.1 KB631741 ERL85707.1 GDIQ01230623 JAK21102.1 GDIQ01031170 JAN63567.1 ATLV01014699 KE524979 KFB39408.1 GBBM01003409 JAC32009.1 GDIP01085727 LRGB01000996 JAM17988.1 KZS14237.1 GDIP01123316 JAL80398.1 GDIP01152337 JAJ71065.1 GDIQ01151868 JAK99857.1 UFQS01000998 UFQT01000998 SSX08358.1 SSX28381.1 MUJZ01057625 OTF72154.1 GDIP01004968 JAM98747.1 GDIQ01032400 JAN62337.1 GDIP01070884 JAM32831.1 GDIQ01093819 JAL57907.1 APGK01040046 APGK01040047 APGK01040048 APGK01040049 APGK01040050 KB740975 ENN76525.1 GDIP01044930 JAM58785.1 GDIP01063003 JAM40712.1 GDIP01072181 JAM31534.1 GDIQ01006671 JAN88066.1 GBBK01001567 JAC22915.1 AXCM01017532 PYGN01000051 PSN56128.1 KK114184 KFM61840.1 PZQS01000004 PVD32493.1 GDIQ01107425 JAL44301.1 CAEY01000116 GFAC01005164 JAT94024.1 NWSH01001233 PCG72021.1 PCG72022.1
GGMR01006175 MBY18794.1 GGMS01004721 MBY73924.1 GBHO01017815 JAG25789.1 GBHO01017816 JAG25788.1 LJIG01001161 KRT85779.1 GDIQ01248423 JAK03302.1 GDIQ01205639 JAK46086.1 GDIQ01249975 JAK01750.1 GDIP01232326 JAI91075.1 GDIQ01107418 GDIP01059485 JAL44308.1 JAM44230.1 GDIP01183816 JAJ39586.1 GDIQ01251468 JAK00257.1 GL732533 EFX84931.1 GDIQ01068031 JAN26706.1 GDIQ01089024 JAN05713.1 GDIQ01049952 JAN44785.1 GDIP01094347 JAM09368.1 GDIQ01144903 JAL06823.1 GGMS01004371 MBY73574.1 NEVH01001335 PNF43116.1 GDIP01185110 JAJ38292.1 AJVK01021811 GDIQ01174096 JAK77629.1 GDIQ01112920 GDIP01085480 LRGB01002384 JAL38806.1 JAM18235.1 KZS07935.1 GFXV01005625 MBW17430.1 ABLF02040673 ABLF02040674 ABLF02040675 ABLF02040682 ABLF02040683 ABLF02040686 ABLF02040688 ABLF02040693 GDIP01165885 JAJ57517.1 GDIP01073234 JAM30481.1 GDIQ01048466 JAN46271.1 GECU01004560 JAT03147.1 KK852544 KDR21624.1 GDIQ01139812 JAL11914.1 GDIP01226307 JAI97094.1 GDIP01220166 JAJ03236.1 GDIQ01176393 JAK75332.1 GDIQ01009264 JAN85473.1 GDIQ01048465 JAN46272.1 GL732530 EFX86305.1 GDIP01218581 JAJ04821.1 APCN01002460 GDIP01199931 JAJ23471.1 GDIP01240121 JAI83280.1 GL768069 EFZ12524.1 AAAB01008964 EAA43883.4 GDIP01247494 JAI75907.1 GDIQ01160111 JAK91614.1 GDIQ01161666 JAK90059.1 GDIQ01137465 JAL14261.1 AMQM01010917 KB095994 ESO09179.1 GEZM01035706 GEZM01035705 JAV83119.1 GDIP01204709 JAJ18693.1 KB631741 ERL85707.1 GDIQ01230623 JAK21102.1 GDIQ01031170 JAN63567.1 ATLV01014699 KE524979 KFB39408.1 GBBM01003409 JAC32009.1 GDIP01085727 LRGB01000996 JAM17988.1 KZS14237.1 GDIP01123316 JAL80398.1 GDIP01152337 JAJ71065.1 GDIQ01151868 JAK99857.1 UFQS01000998 UFQT01000998 SSX08358.1 SSX28381.1 MUJZ01057625 OTF72154.1 GDIP01004968 JAM98747.1 GDIQ01032400 JAN62337.1 GDIP01070884 JAM32831.1 GDIQ01093819 JAL57907.1 APGK01040046 APGK01040047 APGK01040048 APGK01040049 APGK01040050 KB740975 ENN76525.1 GDIP01044930 JAM58785.1 GDIP01063003 JAM40712.1 GDIP01072181 JAM31534.1 GDIQ01006671 JAN88066.1 GBBK01001567 JAC22915.1 AXCM01017532 PYGN01000051 PSN56128.1 KK114184 KFM61840.1 PZQS01000004 PVD32493.1 GDIQ01107425 JAL44301.1 CAEY01000116 GFAC01005164 JAT94024.1 NWSH01001233 PCG72021.1 PCG72022.1
Proteomes
UP000053268
UP000053240
UP000075881
UP000075880
UP000000305
UP000075882
+ More
UP000235965 UP000092462 UP000076858 UP000007819 UP000027135 UP000075885 UP000075840 UP000076407 UP000075901 UP000007062 UP000015101 UP000075900 UP000030742 UP000030765 UP000076408 UP000069272 UP000019118 UP000075884 UP000075883 UP000245037 UP000054359 UP000245119 UP000015104 UP000218220
UP000235965 UP000092462 UP000076858 UP000007819 UP000027135 UP000075885 UP000075840 UP000076407 UP000075901 UP000007062 UP000015101 UP000075900 UP000030742 UP000030765 UP000076408 UP000069272 UP000019118 UP000075884 UP000075883 UP000245037 UP000054359 UP000245119 UP000015104 UP000218220
Interpro
SUPFAM
SSF54695
SSF54695
Gene 3D
ProteinModelPortal
A0A2W1C080
A0A194Q339
A0A0N0PCM8
A0A2S2NNR1
A0A182KDV1
A0A2S2Q878
+ More
A0A0A9Y0S3 A0A0A9Y3U7 A0A0T6BEL8 A0A0P5GGB7 A0A0P5KSL0 A0A0P5FU90 A0A0P4Y7U0 A0A182JLR5 A0A0P5RN71 A0A0P5BJ70 A0A0P5G9Y4 E9G6G5 A0A182L169 A0A0P6ELD9 A0A0P6CNE0 A0A0P6FHY7 A0A0P5VYS4 A0A0P5PSS9 A0A2S2Q771 A0A2J7RQK8 A0A0P5BSD2 A0A1B0D0Z9 A0A0P5MTL3 A0A0P5WJQ2 A0A2H8TU10 J9KBA4 A0A0P5CVD5 A0A0P5YDY4 A0A0P6FFB6 A0A1B6JVD2 A0A067RPK6 A0A0P5PD88 A0A0P4YND9 A0A0P4Z2A9 A0A0P5L222 A0A0P6JTA4 A0A0P6G807 E9G2I2 A0A182PPR6 A0A0P4Z728 A0A182HUF7 A0A182XKZ8 A0A182SFK4 A0A0P5ARG3 A0A0N7ZJ77 E9J3X3 Q7PJ62 A0A182L172 A0A0P4X5J6 A0A0P5M6G9 A0A0P5MM19 A0A0P5PAC5 T1G8N9 A0A182R4S3 A0A1Y1MBB0 A0A0P5ABL6 U4TY14 A0A0P5H9G8 A0A0P6IH24 A0A084VN64 A0A023GDU2 A0A1S4GXL1 A0A0N8CYR5 A0A182Y3M5 A0A0P5TWV6 A0A0N8AFK0 A0A0P5N395 A0A336KTB5 A0A182F5V3 A0A1Y3AWI0 A0A0P6CAE6 A0A0P6GJM9 A0A0P5YJA6 A0A0N8CCA5 N6U4K5 A0A0P5ZEA9 A0A0P5Z2I1 A0A0P5XHT2 A0A2Y9D1K8 A0A0P6IC74 A0A023FQ08 A0A182M9X6 A0A2P8ZHZ7 A0A087T9Q1 A0A2T7PGE8 A0A182F5U9 A0A182K7C2 A0A0P5R5I5 A0A182PPR2 T1KJ18 A0A1E1X451 A0A2A4JJ74 A0A182XKZ3 A0A2A4JKU0
A0A0A9Y0S3 A0A0A9Y3U7 A0A0T6BEL8 A0A0P5GGB7 A0A0P5KSL0 A0A0P5FU90 A0A0P4Y7U0 A0A182JLR5 A0A0P5RN71 A0A0P5BJ70 A0A0P5G9Y4 E9G6G5 A0A182L169 A0A0P6ELD9 A0A0P6CNE0 A0A0P6FHY7 A0A0P5VYS4 A0A0P5PSS9 A0A2S2Q771 A0A2J7RQK8 A0A0P5BSD2 A0A1B0D0Z9 A0A0P5MTL3 A0A0P5WJQ2 A0A2H8TU10 J9KBA4 A0A0P5CVD5 A0A0P5YDY4 A0A0P6FFB6 A0A1B6JVD2 A0A067RPK6 A0A0P5PD88 A0A0P4YND9 A0A0P4Z2A9 A0A0P5L222 A0A0P6JTA4 A0A0P6G807 E9G2I2 A0A182PPR6 A0A0P4Z728 A0A182HUF7 A0A182XKZ8 A0A182SFK4 A0A0P5ARG3 A0A0N7ZJ77 E9J3X3 Q7PJ62 A0A182L172 A0A0P4X5J6 A0A0P5M6G9 A0A0P5MM19 A0A0P5PAC5 T1G8N9 A0A182R4S3 A0A1Y1MBB0 A0A0P5ABL6 U4TY14 A0A0P5H9G8 A0A0P6IH24 A0A084VN64 A0A023GDU2 A0A1S4GXL1 A0A0N8CYR5 A0A182Y3M5 A0A0P5TWV6 A0A0N8AFK0 A0A0P5N395 A0A336KTB5 A0A182F5V3 A0A1Y3AWI0 A0A0P6CAE6 A0A0P6GJM9 A0A0P5YJA6 A0A0N8CCA5 N6U4K5 A0A0P5ZEA9 A0A0P5Z2I1 A0A0P5XHT2 A0A2Y9D1K8 A0A0P6IC74 A0A023FQ08 A0A182M9X6 A0A2P8ZHZ7 A0A087T9Q1 A0A2T7PGE8 A0A182F5U9 A0A182K7C2 A0A0P5R5I5 A0A182PPR2 T1KJ18 A0A1E1X451 A0A2A4JJ74 A0A182XKZ3 A0A2A4JKU0
PDB
6EBM
E-value=4.32885e-35,
Score=365
Ontologies
GO
PANTHER
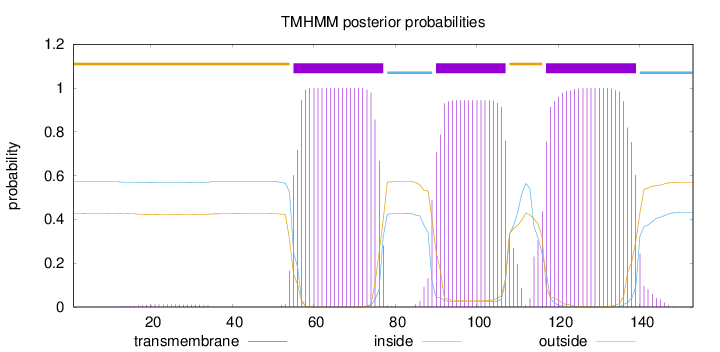
Topology
Length:
153
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
62.61368
Exp number, first 60 AAs:
5.64378
Total prob of N-in:
0.57234
outside
1 - 54
TMhelix
55 - 77
inside
78 - 89
TMhelix
90 - 107
outside
108 - 116
TMhelix
117 - 139
inside
140 - 153
Population Genetic Test Statistics
Pi
292.773916
Theta
177.901259
Tajima's D
2.538861
CLR
0.244626
CSRT
0.949952502374881
Interpretation
Uncertain
