Gene
KWMTBOMO01553
Pre Gene Modal
BGIBMGA009080
Annotation
PREDICTED:_potassium_voltage-gated_channel_protein_Shaw-like_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.715
Sequence
CDS
ATGGACTTTCTCTTGAGCATTTATGAACGAATTCATAATAGATACGAGACTTACAAGGCGACCTTGAAGAAGATCCCCGCGACGCGACTGTCCCGGCTGACGGAGGCGCTCGCCAACTACGACCCGGTGCTCAACGAGTATTTCTTCGACCGGCACCCAGGGGTCTTTGCCCAAGTCCTCAACTACTACAGAACGGGAAAGCTACATTATCCTACGAATGTGTGCGGTCCACTCTTTGAAGAGGAGCTGGAGTTCTGGGGGCTGGACTCCAACCAGGTGGAGCCATGCTGCTGGTCTACTTACAGCATACACAGGGACACGCAAGTAAGTCCTCGTGTCGTGGTTTTATTTACTATACACACGCAAACAGGATCACGTGTCGTTTTGCGTTGGTAG
Protein
MDFLLSIYERIHNRYETYKATLKKIPATRLSRLTEALANYDPVLNEYFFDRHPGVFAQVLNYYRTGKLHYPTNVCGPLFEEELEFWGLDSNQVEPCCWSTYSIHRDTQVSPRVVVLFTIHTQTGSRVVLRW
Summary
Similarity
Belongs to the potassium channel family.
Uniprot
A0A182L169
A0A182M9D1
A0A1S3DKL7
A0A182KEV1
A0A182IZF4
A0A182TAG7
+ More
W5JET9 A0A1B0CZX0 A0A182U897 A0A087T9Q0 T1J6X3 A0A1W4VVD0 A0A2J7RQK8 D6WY23 A0A2H8TU10 A0A182XKZ8 A0A067RPK6 A0A182PPR6 J9KBA4 A0A182F5V3 A0A182W6V5 V4A7E7 A0A034V1N1 A0A1Y1MBB0 A0A182R4S3 A0A2P6KPH2 W8CBV5 A0A182HUF7 A0A084VN64 T1EID3 A0A1J1HPA2 A0A1S4GXL1 A0A2H1CFG2 A0A182QY72 A0A2Y9D1K8 A0A2C9JX97 A0A182Y3M5 A0A0L8GYK9 A0A182L170 A0A0J7KM26 B4KIP8 A0A2T7PGG1 A0A1I8PJ94 A0A132AE88 B5DH77 B3MUT9 B4NY30 A0A1C9TA82 A0A0J9QZ87 A0A1C9TA95 B4JPW4 A0A182GRV4 A0A0M3QWN6 A0A0Q9XE33 A0A1W0XEG0 B4M9S4 A0A1I8N3L4 A0A0L0C8F9 A0A1A9VZZ9 A0A1B0B7I1 A0A0P8XHG9 Q2MHH6 A0A1A9YAT2 A0A1B0A4W8 A8DYR5 B4N1E1 A0A0R3NQJ1 R9PY39 A0A0R1DLX9 A0A0N8NZG3 A0A0R1DL00 A0A183PFG6 A0A0J9R019 A0A0Q5WAL6 A0A0Q5WH32 B3N7Z2 A0A1B0FLL1 A0A0J9TJF9 A0A0R3NQX8 A0A0R1DN84 A0A1Y3AYS1 A0A1B0CZX1 L7MLV5 A0A1S8WJI7 A0A1B6DCB7 A0A2S2Q3L0 E9G6G5 N6T8H1 Q7PNA8 A0A131YDY6 E5SNM7 B7QM66 A0A131YH86 A0A182QU70 T1GRP9 A0A131XME8 A0A2J7RQJ8 A0A084VFD8 A0A1C9TA99 A0A1D2MY37
W5JET9 A0A1B0CZX0 A0A182U897 A0A087T9Q0 T1J6X3 A0A1W4VVD0 A0A2J7RQK8 D6WY23 A0A2H8TU10 A0A182XKZ8 A0A067RPK6 A0A182PPR6 J9KBA4 A0A182F5V3 A0A182W6V5 V4A7E7 A0A034V1N1 A0A1Y1MBB0 A0A182R4S3 A0A2P6KPH2 W8CBV5 A0A182HUF7 A0A084VN64 T1EID3 A0A1J1HPA2 A0A1S4GXL1 A0A2H1CFG2 A0A182QY72 A0A2Y9D1K8 A0A2C9JX97 A0A182Y3M5 A0A0L8GYK9 A0A182L170 A0A0J7KM26 B4KIP8 A0A2T7PGG1 A0A1I8PJ94 A0A132AE88 B5DH77 B3MUT9 B4NY30 A0A1C9TA82 A0A0J9QZ87 A0A1C9TA95 B4JPW4 A0A182GRV4 A0A0M3QWN6 A0A0Q9XE33 A0A1W0XEG0 B4M9S4 A0A1I8N3L4 A0A0L0C8F9 A0A1A9VZZ9 A0A1B0B7I1 A0A0P8XHG9 Q2MHH6 A0A1A9YAT2 A0A1B0A4W8 A8DYR5 B4N1E1 A0A0R3NQJ1 R9PY39 A0A0R1DLX9 A0A0N8NZG3 A0A0R1DL00 A0A183PFG6 A0A0J9R019 A0A0Q5WAL6 A0A0Q5WH32 B3N7Z2 A0A1B0FLL1 A0A0J9TJF9 A0A0R3NQX8 A0A0R1DN84 A0A1Y3AYS1 A0A1B0CZX1 L7MLV5 A0A1S8WJI7 A0A1B6DCB7 A0A2S2Q3L0 E9G6G5 N6T8H1 Q7PNA8 A0A131YDY6 E5SNM7 B7QM66 A0A131YH86 A0A182QU70 T1GRP9 A0A131XME8 A0A2J7RQJ8 A0A084VFD8 A0A1C9TA99 A0A1D2MY37
Pubmed
20966253
20920257
23761445
18362917
19820115
24845553
+ More
23254933 25348373 28004739 24495485 24438588 12364791 15562597 25244985 17994087 26555130 15632085 17550304 22936249 26483478 25315136 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23185243 25576852 21292972 23537049 26830274 28049606 27289101
23254933 25348373 28004739 24495485 24438588 12364791 15562597 25244985 17994087 26555130 15632085 17550304 22936249 26483478 25315136 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23185243 25576852 21292972 23537049 26830274 28049606 27289101
EMBL
AXCM01016303
ADMH02001711
ETN61339.1
AJVK01009752
KK114184
KFM61839.1
+ More
JH431896 NEVH01001335 PNF43116.1 KQ971362 EFA08935.1 GFXV01005625 MBW17430.1 KK852544 KDR21624.1 ABLF02040673 ABLF02040674 ABLF02040675 ABLF02040682 ABLF02040683 ABLF02040686 ABLF02040688 ABLF02040693 KB202047 ESO92677.1 GAKP01022897 JAC36057.1 GEZM01035706 GEZM01035705 JAV83119.1 MWRG01007515 PRD28222.1 GAMC01004476 JAC02080.1 APCN01002460 ATLV01014699 KE524979 KFB39408.1 AMQM01005141 AMQM01005142 KB096830 ESO01058.1 CVRI01000014 CRK89787.1 AAAB01008964 KZ429578 PIS86208.1 AXCN02000817 KQ419930 KOF81934.1 LBMM01005697 KMQ91286.1 CH933807 EDW12404.2 PZQS01000004 PVD32494.1 JXLN01013212 KPM09169.1 CH379058 EDY69652.2 CH902624 EDV33004.2 CM000157 EDW88632.2 KU681444 AOR07224.1 CM002910 KMY89248.1 KU681455 AOR07236.1 CH916372 EDV98944.1 JXUM01083298 KQ563367 KXJ73970.1 CP012525 ALC44463.1 KRG03236.1 MTYJ01000001 OQV25870.1 CH940654 EDW57950.2 JRES01000864 KNC27689.1 JXJN01009667 KPU74245.1 AB212763 BAE75955.1 AE014134 ABV53658.2 CH963920 EDW78078.2 KRT03384.1 KRT03385.1 AAF52768.4 KRJ97950.1 KPU74244.1 KRJ97951.1 UZAL01033156 VDP62733.1 KMY89249.1 CH954177 KQS70372.1 KQS70373.1 EDV58353.2 CCAG010008497 KMY89250.1 KRT03386.1 KRJ97949.1 MUJZ01053887 OTF72954.1 AJVK01009762 AJVK01009763 AJVK01009764 AJVK01009765 GACK01000192 JAA64842.1 KV906658 OON14423.1 GEDC01014043 JAS23255.1 GGMS01002609 MBY71812.1 GL732533 EFX84931.1 APGK01040038 APGK01040039 APGK01040040 KB740975 ENN76524.1 EAA12472.5 GEDV01012426 JAP76131.1 ABJB010403212 DS969391 EEC19938.1 GEDV01009948 JAP78609.1 AXCN02000818 CAQQ02032997 GEFH01001273 JAP67308.1 PNF43105.1 ATLV01012379 KE524786 KFB36682.1 KU681456 AOR07237.1 LJIJ01000397 ODM97910.1
JH431896 NEVH01001335 PNF43116.1 KQ971362 EFA08935.1 GFXV01005625 MBW17430.1 KK852544 KDR21624.1 ABLF02040673 ABLF02040674 ABLF02040675 ABLF02040682 ABLF02040683 ABLF02040686 ABLF02040688 ABLF02040693 KB202047 ESO92677.1 GAKP01022897 JAC36057.1 GEZM01035706 GEZM01035705 JAV83119.1 MWRG01007515 PRD28222.1 GAMC01004476 JAC02080.1 APCN01002460 ATLV01014699 KE524979 KFB39408.1 AMQM01005141 AMQM01005142 KB096830 ESO01058.1 CVRI01000014 CRK89787.1 AAAB01008964 KZ429578 PIS86208.1 AXCN02000817 KQ419930 KOF81934.1 LBMM01005697 KMQ91286.1 CH933807 EDW12404.2 PZQS01000004 PVD32494.1 JXLN01013212 KPM09169.1 CH379058 EDY69652.2 CH902624 EDV33004.2 CM000157 EDW88632.2 KU681444 AOR07224.1 CM002910 KMY89248.1 KU681455 AOR07236.1 CH916372 EDV98944.1 JXUM01083298 KQ563367 KXJ73970.1 CP012525 ALC44463.1 KRG03236.1 MTYJ01000001 OQV25870.1 CH940654 EDW57950.2 JRES01000864 KNC27689.1 JXJN01009667 KPU74245.1 AB212763 BAE75955.1 AE014134 ABV53658.2 CH963920 EDW78078.2 KRT03384.1 KRT03385.1 AAF52768.4 KRJ97950.1 KPU74244.1 KRJ97951.1 UZAL01033156 VDP62733.1 KMY89249.1 CH954177 KQS70372.1 KQS70373.1 EDV58353.2 CCAG010008497 KMY89250.1 KRT03386.1 KRJ97949.1 MUJZ01053887 OTF72954.1 AJVK01009762 AJVK01009763 AJVK01009764 AJVK01009765 GACK01000192 JAA64842.1 KV906658 OON14423.1 GEDC01014043 JAS23255.1 GGMS01002609 MBY71812.1 GL732533 EFX84931.1 APGK01040038 APGK01040039 APGK01040040 KB740975 ENN76524.1 EAA12472.5 GEDV01012426 JAP76131.1 ABJB010403212 DS969391 EEC19938.1 GEDV01009948 JAP78609.1 AXCN02000818 CAQQ02032997 GEFH01001273 JAP67308.1 PNF43105.1 ATLV01012379 KE524786 KFB36682.1 KU681456 AOR07237.1 LJIJ01000397 ODM97910.1
Proteomes
UP000075882
UP000075883
UP000079169
UP000075881
UP000075880
UP000075901
+ More
UP000000673 UP000092462 UP000075902 UP000054359 UP000192221 UP000235965 UP000007266 UP000076407 UP000027135 UP000075885 UP000007819 UP000069272 UP000075920 UP000030746 UP000075900 UP000075840 UP000030765 UP000015101 UP000183832 UP000075886 UP000075884 UP000076420 UP000076408 UP000053454 UP000036403 UP000009192 UP000245119 UP000095300 UP000001819 UP000007801 UP000002282 UP000001070 UP000069940 UP000249989 UP000092553 UP000008792 UP000095301 UP000037069 UP000091820 UP000092460 UP000092443 UP000092445 UP000000803 UP000007798 UP000050791 UP000008711 UP000092444 UP000000305 UP000019118 UP000007062 UP000001555 UP000015102 UP000094527
UP000000673 UP000092462 UP000075902 UP000054359 UP000192221 UP000235965 UP000007266 UP000076407 UP000027135 UP000075885 UP000007819 UP000069272 UP000075920 UP000030746 UP000075900 UP000075840 UP000030765 UP000015101 UP000183832 UP000075886 UP000075884 UP000076420 UP000076408 UP000053454 UP000036403 UP000009192 UP000245119 UP000095300 UP000001819 UP000007801 UP000002282 UP000001070 UP000069940 UP000249989 UP000092553 UP000008792 UP000095301 UP000037069 UP000091820 UP000092460 UP000092443 UP000092445 UP000000803 UP000007798 UP000050791 UP000008711 UP000092444 UP000000305 UP000019118 UP000007062 UP000001555 UP000015102 UP000094527
PRIDE
Interpro
SUPFAM
SSF54695
SSF54695
Gene 3D
ProteinModelPortal
A0A182L169
A0A182M9D1
A0A1S3DKL7
A0A182KEV1
A0A182IZF4
A0A182TAG7
+ More
W5JET9 A0A1B0CZX0 A0A182U897 A0A087T9Q0 T1J6X3 A0A1W4VVD0 A0A2J7RQK8 D6WY23 A0A2H8TU10 A0A182XKZ8 A0A067RPK6 A0A182PPR6 J9KBA4 A0A182F5V3 A0A182W6V5 V4A7E7 A0A034V1N1 A0A1Y1MBB0 A0A182R4S3 A0A2P6KPH2 W8CBV5 A0A182HUF7 A0A084VN64 T1EID3 A0A1J1HPA2 A0A1S4GXL1 A0A2H1CFG2 A0A182QY72 A0A2Y9D1K8 A0A2C9JX97 A0A182Y3M5 A0A0L8GYK9 A0A182L170 A0A0J7KM26 B4KIP8 A0A2T7PGG1 A0A1I8PJ94 A0A132AE88 B5DH77 B3MUT9 B4NY30 A0A1C9TA82 A0A0J9QZ87 A0A1C9TA95 B4JPW4 A0A182GRV4 A0A0M3QWN6 A0A0Q9XE33 A0A1W0XEG0 B4M9S4 A0A1I8N3L4 A0A0L0C8F9 A0A1A9VZZ9 A0A1B0B7I1 A0A0P8XHG9 Q2MHH6 A0A1A9YAT2 A0A1B0A4W8 A8DYR5 B4N1E1 A0A0R3NQJ1 R9PY39 A0A0R1DLX9 A0A0N8NZG3 A0A0R1DL00 A0A183PFG6 A0A0J9R019 A0A0Q5WAL6 A0A0Q5WH32 B3N7Z2 A0A1B0FLL1 A0A0J9TJF9 A0A0R3NQX8 A0A0R1DN84 A0A1Y3AYS1 A0A1B0CZX1 L7MLV5 A0A1S8WJI7 A0A1B6DCB7 A0A2S2Q3L0 E9G6G5 N6T8H1 Q7PNA8 A0A131YDY6 E5SNM7 B7QM66 A0A131YH86 A0A182QU70 T1GRP9 A0A131XME8 A0A2J7RQJ8 A0A084VFD8 A0A1C9TA99 A0A1D2MY37
W5JET9 A0A1B0CZX0 A0A182U897 A0A087T9Q0 T1J6X3 A0A1W4VVD0 A0A2J7RQK8 D6WY23 A0A2H8TU10 A0A182XKZ8 A0A067RPK6 A0A182PPR6 J9KBA4 A0A182F5V3 A0A182W6V5 V4A7E7 A0A034V1N1 A0A1Y1MBB0 A0A182R4S3 A0A2P6KPH2 W8CBV5 A0A182HUF7 A0A084VN64 T1EID3 A0A1J1HPA2 A0A1S4GXL1 A0A2H1CFG2 A0A182QY72 A0A2Y9D1K8 A0A2C9JX97 A0A182Y3M5 A0A0L8GYK9 A0A182L170 A0A0J7KM26 B4KIP8 A0A2T7PGG1 A0A1I8PJ94 A0A132AE88 B5DH77 B3MUT9 B4NY30 A0A1C9TA82 A0A0J9QZ87 A0A1C9TA95 B4JPW4 A0A182GRV4 A0A0M3QWN6 A0A0Q9XE33 A0A1W0XEG0 B4M9S4 A0A1I8N3L4 A0A0L0C8F9 A0A1A9VZZ9 A0A1B0B7I1 A0A0P8XHG9 Q2MHH6 A0A1A9YAT2 A0A1B0A4W8 A8DYR5 B4N1E1 A0A0R3NQJ1 R9PY39 A0A0R1DLX9 A0A0N8NZG3 A0A0R1DL00 A0A183PFG6 A0A0J9R019 A0A0Q5WAL6 A0A0Q5WH32 B3N7Z2 A0A1B0FLL1 A0A0J9TJF9 A0A0R3NQX8 A0A0R1DN84 A0A1Y3AYS1 A0A1B0CZX1 L7MLV5 A0A1S8WJI7 A0A1B6DCB7 A0A2S2Q3L0 E9G6G5 N6T8H1 Q7PNA8 A0A131YDY6 E5SNM7 B7QM66 A0A131YH86 A0A182QU70 T1GRP9 A0A131XME8 A0A2J7RQJ8 A0A084VFD8 A0A1C9TA99 A0A1D2MY37
PDB
3KVT
E-value=2.68822e-49,
Score=486
Ontologies
GO
PANTHER
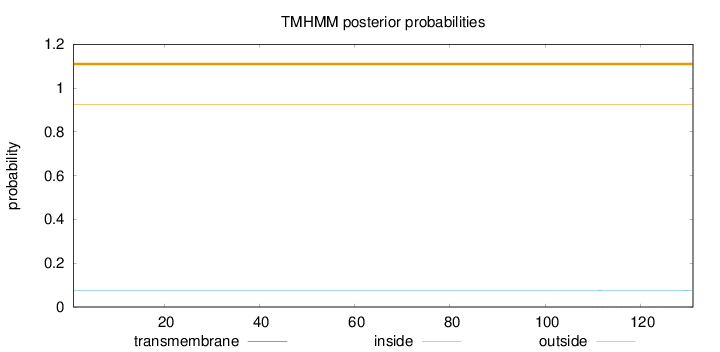
Topology
Length:
131
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01001
Exp number, first 60 AAs:
0.00027
Total prob of N-in:
0.07607
outside
1 - 131
Population Genetic Test Statistics
Pi
235.993218
Theta
158.257753
Tajima's D
1.702054
CLR
0.098135
CSRT
0.829758512074396
Interpretation
Uncertain
