Gene
KWMTBOMO01550
Pre Gene Modal
BGIBMGA009074
Annotation
PREDICTED:_uncharacterized_protein_LOC106138832_[Amyelois_transitella]
Full name
tRNA-dihydrouridine(47) synthase [NAD(P)(+)]
Alternative Name
tRNA-dihydrouridine synthase 3
Location in the cell
Nuclear Reliability : 2.359
Sequence
CDS
ATGAGAATAACAGATGTAAAATCGGAAAAACTCATAGAGGCCGTTAAGAAGAGACCGGCGTTGTATAATAAAAGTGACACTATGTATTACTGTCAGAAAAAGAACAAAATCAAATTGTGGATCGAAGTCTGCCAAGAAGTGTTCAGTAATTGGGAGCAAATGTCAGCACACGAAAAAGTTGAACTGCGCCACGAAATACTAAAACGCTGGAAGAGCCTCCGAACGTGCTTCTCAAGGGAACTATCGTTACAGAAGAAAGAGAAACGCAGCCTCGAAATCGATCATGAACCGATTAAAAGACGTAAAAAGTACGAATACTTTGGTTTGATGTCCTTTTTACTTCACCCGGACGCAATAGGTGAATTCGAAATAACGAAAGAGACAGACGAAGAGTCCTCTGATCCTTTGGATTGTATGAAAACTGAGATTTACGACGAGGGACAAACATCTAAAGTAGAAATAGTTAATGAAAGCGTCAACAATCAGTTTCAACCGTTCTACGAACGAAATGAAAACGTTGACGAGAAGATTCTGGAAATATTAAAAGAAATGAAGAAAGATGAATGCGACGAGGACAGACAGTTTATGTTGTCATTAGTTCCAAGCTTTCGGAAATTAAATGATAAGCAGAAGTTTGAAGCTAGAATAGGAATGTTAAAGGTCCTACAAGAGATAACGTTTCAGGAGAAAGAGACTCTATGA
Protein
MRITDVKSEKLIEAVKKRPALYNKSDTMYYCQKKNKIKLWIEVCQEVFSNWEQMSAHEKVELRHEILKRWKSLRTCFSRELSLQKKEKRSLEIDHEPIKRRKKYEYFGLMSFLLHPDAIGEFEITKETDEESSDPLDCMKTEIYDEGQTSKVEIVNESVNNQFQPFYERNENVDEKILEILKEMKKDECDEDRQFMLSLVPSFRKLNDKQKFEARIGMLKVLQEITFQEKETL
Summary
Cofactor
FMN
Similarity
Belongs to the dus family. Dus3 subfamily.
Uniprot
H9JHS6
A0A2A4IZR8
A0A2H1W8X3
A0A067RA73
A0A2S2NEL4
A0A1L8FUL7
+ More
A0A067QVH3 A0A1B6DML8 A0A194QNL3 A0A0N0BFW2 A0A0L7R227 A0A026WD33 D6WRU3 F4WMC5 A0A158P282 A0A195BR61 A0A195F3Q1 A0A151XC39 A0A2S2QKM5 A0A0J7L3L1 A0A212F8H6 A0A310SQK1 A0A212FBS8 A0A195DC77 A0A2A3ERT0 A0A088AEC2 J9KU42 A0A2S2PS33 A0A151IFB9 A0A154P6K1 A0A0P4W9X1 A0A0P4W354 E2AWC0 S4P5P6 A0A0N5D5I8 A0A194QLR2 A0A1B6JUW1 A0A194QMN5 A0A2A4J5L3 A0A2H1WCZ7 A0A194Q210 A0A1E1WAP8 A0A1I8QBW5 A0A1E1W082 A0A2A4K4Y9 A0A0N0PC80 A0A0P4WL80 J9EDY0 A0A0L0BVE8 A0A3P7E1U8 N6U963 A0A2W1BS85 A0A067QMQ0 A0A0R3R2M1 A0A0H5S1L7 A0A2H1X0E4 A0A0A9WF21 A0A212FPD0 A0A1E1WMV5 A0A044V964 A0A3P6U100 A0A182EJB9 A0A0N4TX01 A0A1B6CC07 A0A2A4K541 A0A2H1WP32 A0A2H1WG94 A0A0L7KQU0 A0A2H1WWQ0 A0A146KXI5 A0A0A9XAN0 A0A212F723 A0A0A9X5S0
A0A067QVH3 A0A1B6DML8 A0A194QNL3 A0A0N0BFW2 A0A0L7R227 A0A026WD33 D6WRU3 F4WMC5 A0A158P282 A0A195BR61 A0A195F3Q1 A0A151XC39 A0A2S2QKM5 A0A0J7L3L1 A0A212F8H6 A0A310SQK1 A0A212FBS8 A0A195DC77 A0A2A3ERT0 A0A088AEC2 J9KU42 A0A2S2PS33 A0A151IFB9 A0A154P6K1 A0A0P4W9X1 A0A0P4W354 E2AWC0 S4P5P6 A0A0N5D5I8 A0A194QLR2 A0A1B6JUW1 A0A194QMN5 A0A2A4J5L3 A0A2H1WCZ7 A0A194Q210 A0A1E1WAP8 A0A1I8QBW5 A0A1E1W082 A0A2A4K4Y9 A0A0N0PC80 A0A0P4WL80 J9EDY0 A0A0L0BVE8 A0A3P7E1U8 N6U963 A0A2W1BS85 A0A067QMQ0 A0A0R3R2M1 A0A0H5S1L7 A0A2H1X0E4 A0A0A9WF21 A0A212FPD0 A0A1E1WMV5 A0A044V964 A0A3P6U100 A0A182EJB9 A0A0N4TX01 A0A1B6CC07 A0A2A4K541 A0A2H1WP32 A0A2H1WG94 A0A0L7KQU0 A0A2H1WWQ0 A0A146KXI5 A0A0A9XAN0 A0A212F723 A0A0A9X5S0
EC Number
1.3.1.-
Pubmed
EMBL
BABH01029498
NWSH01004566
PCG64918.1
ODYU01007093
SOQ49555.1
KK852602
+ More
KDR20478.1 GGMR01002773 MBY15392.1 CM004476 OCT75290.1 KK853328 KDR08446.1 GEDC01010406 JAS26892.1 KQ461191 KPJ07113.1 KQ435794 KOX73672.1 KQ414666 KOC64878.1 KK107261 QOIP01000012 EZA53972.1 RLU16330.1 KQ971351 EFA06435.1 GL888217 EGI64769.1 ADTU01001164 KQ976418 KYM89451.1 KQ981855 KYN34807.1 KQ982315 KYQ57858.1 GGMS01009116 MBY78319.1 LBMM01000937 KMQ97188.1 AGBW02009720 OWR50045.1 KQ761662 OAD57183.1 AGBW02009279 OWR51189.1 KQ980989 KYN10515.1 KZ288192 PBC34400.1 ABLF02030014 GGMR01019594 MBY32213.1 KQ977799 KYM99596.1 KQ434827 KZC07471.1 GDRN01106646 GDRN01106645 JAI57540.1 GDRN01106643 JAI57541.1 GL443280 EFN62280.1 GAIX01008457 JAA84103.1 UYYF01004606 VDN05797.1 KQ461196 KPJ06467.1 GECU01004715 JAT02992.1 KQ458860 KPJ04746.1 NWSH01003192 PCG66814.1 ODYU01007817 SOQ50907.1 KQ459582 KPI99034.1 GDQN01006974 JAT84080.1 GDQN01010705 JAT80349.1 NWSH01000176 PCG78720.1 KQ460651 KPJ13044.1 GDRN01021545 JAI67801.1 ADBV01004300 EJW80661.1 JRES01001388 KNC23209.1 UYWW01001533 VDM10647.1 APGK01035269 KB740923 ENN78240.1 KZ149935 PZC77191.1 KK853142 KDR10746.1 UZAG01019009 VDO41994.1 LN856534 CRZ22603.1 ODYU01012021 SOQ58104.1 GBHO01040141 GBHO01040140 GBHO01040139 GDHC01012787 GDHC01010302 GDHC01004453 JAG03463.1 JAG03464.1 JAG03465.1 JAQ05842.1 JAQ08327.1 JAQ14176.1 AGBW02003328 OWR55560.1 GDQN01002878 JAT88176.1 CMVM020000255 UYRW01003313 VDK88653.1 UZAD01013389 VDN94591.1 GEDC01026463 JAS10835.1 NWSH01000149 PCG79034.1 ODYU01010020 SOQ54829.1 ODYU01008462 SOQ52093.1 JTDY01007070 KOB65480.1 ODYU01011631 SOQ57499.1 GDHC01019159 GDHC01006188 JAP99469.1 JAQ12441.1 GBHO01029489 JAG14115.1 AGBW02009942 OWR49530.1 GBHO01029486 JAG14118.1
KDR20478.1 GGMR01002773 MBY15392.1 CM004476 OCT75290.1 KK853328 KDR08446.1 GEDC01010406 JAS26892.1 KQ461191 KPJ07113.1 KQ435794 KOX73672.1 KQ414666 KOC64878.1 KK107261 QOIP01000012 EZA53972.1 RLU16330.1 KQ971351 EFA06435.1 GL888217 EGI64769.1 ADTU01001164 KQ976418 KYM89451.1 KQ981855 KYN34807.1 KQ982315 KYQ57858.1 GGMS01009116 MBY78319.1 LBMM01000937 KMQ97188.1 AGBW02009720 OWR50045.1 KQ761662 OAD57183.1 AGBW02009279 OWR51189.1 KQ980989 KYN10515.1 KZ288192 PBC34400.1 ABLF02030014 GGMR01019594 MBY32213.1 KQ977799 KYM99596.1 KQ434827 KZC07471.1 GDRN01106646 GDRN01106645 JAI57540.1 GDRN01106643 JAI57541.1 GL443280 EFN62280.1 GAIX01008457 JAA84103.1 UYYF01004606 VDN05797.1 KQ461196 KPJ06467.1 GECU01004715 JAT02992.1 KQ458860 KPJ04746.1 NWSH01003192 PCG66814.1 ODYU01007817 SOQ50907.1 KQ459582 KPI99034.1 GDQN01006974 JAT84080.1 GDQN01010705 JAT80349.1 NWSH01000176 PCG78720.1 KQ460651 KPJ13044.1 GDRN01021545 JAI67801.1 ADBV01004300 EJW80661.1 JRES01001388 KNC23209.1 UYWW01001533 VDM10647.1 APGK01035269 KB740923 ENN78240.1 KZ149935 PZC77191.1 KK853142 KDR10746.1 UZAG01019009 VDO41994.1 LN856534 CRZ22603.1 ODYU01012021 SOQ58104.1 GBHO01040141 GBHO01040140 GBHO01040139 GDHC01012787 GDHC01010302 GDHC01004453 JAG03463.1 JAG03464.1 JAG03465.1 JAQ05842.1 JAQ08327.1 JAQ14176.1 AGBW02003328 OWR55560.1 GDQN01002878 JAT88176.1 CMVM020000255 UYRW01003313 VDK88653.1 UZAD01013389 VDN94591.1 GEDC01026463 JAS10835.1 NWSH01000149 PCG79034.1 ODYU01010020 SOQ54829.1 ODYU01008462 SOQ52093.1 JTDY01007070 KOB65480.1 ODYU01011631 SOQ57499.1 GDHC01019159 GDHC01006188 JAP99469.1 JAQ12441.1 GBHO01029489 JAG14115.1 AGBW02009942 OWR49530.1 GBHO01029486 JAG14118.1
Proteomes
UP000005204
UP000218220
UP000027135
UP000186698
UP000053240
UP000053105
+ More
UP000053825 UP000053097 UP000279307 UP000007266 UP000007755 UP000005205 UP000078540 UP000078541 UP000075809 UP000036403 UP000007151 UP000078492 UP000242457 UP000005203 UP000007819 UP000078542 UP000076502 UP000000311 UP000046394 UP000276776 UP000053268 UP000095300 UP000004810 UP000093561 UP000037069 UP000270924 UP000019118 UP000050602 UP000006672 UP000024404 UP000271087 UP000077448 UP000038020 UP000278627 UP000037510
UP000053825 UP000053097 UP000279307 UP000007266 UP000007755 UP000005205 UP000078540 UP000078541 UP000075809 UP000036403 UP000007151 UP000078492 UP000242457 UP000005203 UP000007819 UP000078542 UP000076502 UP000000311 UP000046394 UP000276776 UP000053268 UP000095300 UP000004810 UP000093561 UP000037069 UP000270924 UP000019118 UP000050602 UP000006672 UP000024404 UP000271087 UP000077448 UP000038020 UP000278627 UP000037510
PRIDE
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
H9JHS6
A0A2A4IZR8
A0A2H1W8X3
A0A067RA73
A0A2S2NEL4
A0A1L8FUL7
+ More
A0A067QVH3 A0A1B6DML8 A0A194QNL3 A0A0N0BFW2 A0A0L7R227 A0A026WD33 D6WRU3 F4WMC5 A0A158P282 A0A195BR61 A0A195F3Q1 A0A151XC39 A0A2S2QKM5 A0A0J7L3L1 A0A212F8H6 A0A310SQK1 A0A212FBS8 A0A195DC77 A0A2A3ERT0 A0A088AEC2 J9KU42 A0A2S2PS33 A0A151IFB9 A0A154P6K1 A0A0P4W9X1 A0A0P4W354 E2AWC0 S4P5P6 A0A0N5D5I8 A0A194QLR2 A0A1B6JUW1 A0A194QMN5 A0A2A4J5L3 A0A2H1WCZ7 A0A194Q210 A0A1E1WAP8 A0A1I8QBW5 A0A1E1W082 A0A2A4K4Y9 A0A0N0PC80 A0A0P4WL80 J9EDY0 A0A0L0BVE8 A0A3P7E1U8 N6U963 A0A2W1BS85 A0A067QMQ0 A0A0R3R2M1 A0A0H5S1L7 A0A2H1X0E4 A0A0A9WF21 A0A212FPD0 A0A1E1WMV5 A0A044V964 A0A3P6U100 A0A182EJB9 A0A0N4TX01 A0A1B6CC07 A0A2A4K541 A0A2H1WP32 A0A2H1WG94 A0A0L7KQU0 A0A2H1WWQ0 A0A146KXI5 A0A0A9XAN0 A0A212F723 A0A0A9X5S0
A0A067QVH3 A0A1B6DML8 A0A194QNL3 A0A0N0BFW2 A0A0L7R227 A0A026WD33 D6WRU3 F4WMC5 A0A158P282 A0A195BR61 A0A195F3Q1 A0A151XC39 A0A2S2QKM5 A0A0J7L3L1 A0A212F8H6 A0A310SQK1 A0A212FBS8 A0A195DC77 A0A2A3ERT0 A0A088AEC2 J9KU42 A0A2S2PS33 A0A151IFB9 A0A154P6K1 A0A0P4W9X1 A0A0P4W354 E2AWC0 S4P5P6 A0A0N5D5I8 A0A194QLR2 A0A1B6JUW1 A0A194QMN5 A0A2A4J5L3 A0A2H1WCZ7 A0A194Q210 A0A1E1WAP8 A0A1I8QBW5 A0A1E1W082 A0A2A4K4Y9 A0A0N0PC80 A0A0P4WL80 J9EDY0 A0A0L0BVE8 A0A3P7E1U8 N6U963 A0A2W1BS85 A0A067QMQ0 A0A0R3R2M1 A0A0H5S1L7 A0A2H1X0E4 A0A0A9WF21 A0A212FPD0 A0A1E1WMV5 A0A044V964 A0A3P6U100 A0A182EJB9 A0A0N4TX01 A0A1B6CC07 A0A2A4K541 A0A2H1WP32 A0A2H1WG94 A0A0L7KQU0 A0A2H1WWQ0 A0A146KXI5 A0A0A9XAN0 A0A212F723 A0A0A9X5S0
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
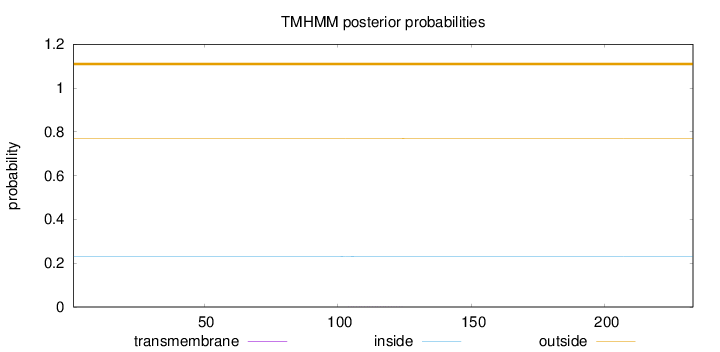
Length:
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00533
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.23144
outside
1 - 233
Population Genetic Test Statistics
Pi
349.087493
Theta
213.545456
Tajima's D
2.040236
CLR
0
CSRT
0.888205589720514
Interpretation
Uncertain
