Gene
KWMTBOMO01545
Pre Gene Modal
BGIBMGA009091
Annotation
PREDICTED:_zonadhesin-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.759
Sequence
CDS
ATGGCCGCCAAACATAACTTCCCCATGCTCCTGCTCGTATCCCTGATGGCTCTCGCAGCCAGCCAGGGTTCGGTTGAAAAAGATTGTCCTGAGAATTCTCATTTGACAATGAACCCCTGCGCGCCGACCTGTGAAGATCCAGACCTGACACACACCAGCTGCGTAGCGGCATTGCTCCCAACATGCCACTGTGATGATGGCTTCTTGTTCGACAAATCTGGAAAATGCGTACCCGTTGATGAATGTCCAGACCAAGAGAACCAATAA
Protein
MAAKHNFPMLLLVSLMALAASQGSVEKDCPENSHLTMNPCAPTCEDPDLTHTSCVAALLPTCHCDDGFLFDKSGKCVPVDECPDQENQ
Summary
Uniprot
EMBL
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
Ontologies
PANTHER
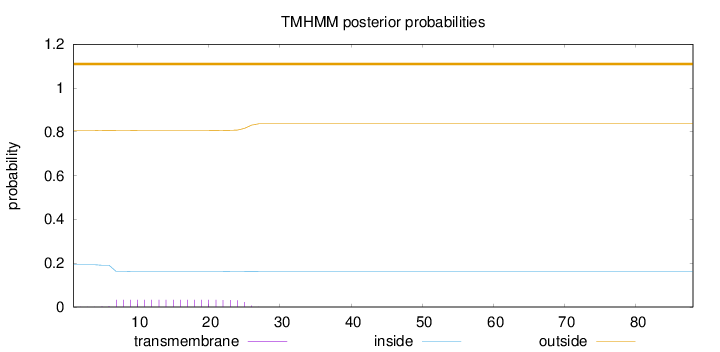
Topology
Subcellular location
Secreted
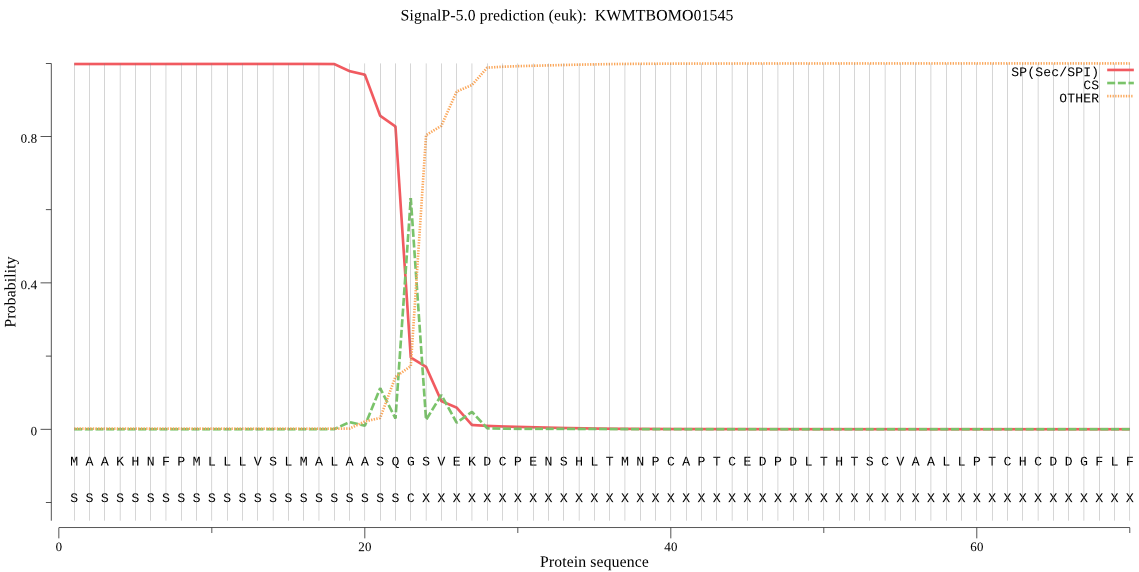
SignalP
Position: 1 - 23,
Likelihood: 0.998283
Length:
88
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.62669
Exp number, first 60 AAs:
0.62669
Total prob of N-in:
0.19327
outside
1 - 88
Population Genetic Test Statistics
Pi
217.870326
Theta
137.838085
Tajima's D
1.958822
CLR
0.485
CSRT
0.87805609719514
Interpretation
Uncertain
