Gene
KWMTBOMO01543 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009092
Annotation
PREDICTED:_zonadhesin-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.093 Nuclear Reliability : 1.702
Sequence
CDS
ATGGCCGCCAAACATTACTTCACCATGCTCCTGCTCGTATCCCTGATGGCTCTCATAGCCAGTAACAGTTCGTTTGAAAAAGATTGTCCTGAGAATTCTCATTTGACAATGGACCCCTGCGCGCCGACCTGTGAAGATCCAGACCTAACACACACCAGCTGCGTAGCGGCATTGCTCCCAACATGCCACTGTGATGATGGCTTCCTCTTCGACAAATCAGGAAAATGCGTACCCGTTGATGAATGTCCAGACCAAAAGAATTGTCCTGAGAATTCTCATTTGACAATGGATCCCTGCGCGCCGACCTGTGAAGACCCGGAACTGAAAAACACCAGCTGCGTAGCGGCATTGCTCCCAACATGCCACTGTGATGATGGCTTCCTCTTCGACAAATCAGGAAAATGCGTACCCGTTGATGAATGTCCAGACCATAAGAATTGTCCTGAGAATTCTCATTTGACAATGGACCCCTGCGCGCCGACCTGTGAAGACCCAGAACTGAAAAACACCAGCTGCGCAGCGGCATTGCTCCCAACATGCCACTGTGATGATGGCTTCCTCTTCGACAAATCAGGAAAATGCGTACCCGTTGAGGAATGTCCAGACCAAAAGAATGAGTGCGTCAACTGA
Protein
MAAKHYFTMLLLVSLMALIASNSSFEKDCPENSHLTMDPCAPTCEDPDLTHTSCVAALLPTCHCDDGFLFDKSGKCVPVDECPDQKNCPENSHLTMDPCAPTCEDPELKNTSCVAALLPTCHCDDGFLFDKSGKCVPVDECPDHKNCPENSHLTMDPCAPTCEDPELKNTSCAAALLPTCHCDDGFLFDKSGKCVPVEECPDQKNECVN
Summary
Similarity
Belongs to the serpin family.
Uniprot
C3ZK78
A0A1J1HWF2
A0A183J2C6
A0A2H1WD28
A0A1J1IUR1
A0A0C2GUB0
+ More
F1LEH5 A0A2H1VKZ4 A0A1J1HI61 A0A2H2IN21 A0A2P6L3P9 A0A0V1MZF6 A0A0V1J3G8 A0A0V1H1U7 A0A087TD78 A0A0V1CXK4 A0A2W1B8Z9 A0A016VC00 E5S2I0 A0A016VAF8 A0A016V9B6 A0A368GXV9 A0A0D6LGI9 A0A090N0V2 A0A1Y1KW09 A0A2P6LAR8 A0A0V1NT84 A0A0V0Z760 A0A0V1LSL0 A0A0V0VIZ3 A0A0V0U5N5 A0A1I8A2H2 A0A0V1B9Z3 A0A0V1B9Y2 A0A0L7LIN3 A0A0V0WZ71 A0A0V1FGY8 A0A0V1J3E7 A0A131ZVD5 A0A194Q2H2 A0A0V0WZ93 A0A368F3L2 A0A0V0Z7C1 A0A0V0Z7M5 A0A087UJV8 A0A1I7Z116 A0A090L0U6 A0A0R3UUA3 A0A1I8A6G1 A0A0V0Z791 A0A0N4Y782 A0A0V0Z793 A0A0N5A6N4 A0A0V0U5E1 A0A194Q8M7 A0A2A4JCJ6 A0A336LED8 A0A0V1EPP9 A0A0K0EAF5 A0A0V0S515 A0A0V1EPP1 H2L2K9 A0A212EIN9 A0A0V1EPN2 A0A2A4J7B9 E4WQ01 D2A397 A0A1I7T4K9 A0A2L2YKC7 H9JHU8 A0A260Z7K7 B0XBB5 H3G2Y3 A0A0V0S563 E4XIX0 A0A2P6L3Q8 A0A0C2D8P6 A0A194QS46 A0A0V0S5K9 A0A1I8BL05 A0A016VB53 A0A2L2XYH3 A0A2L2XYN6 A0A0V0U579 A0A194QS41 C3Y5D2 A0A0C2CJT7 B7P7B7
F1LEH5 A0A2H1VKZ4 A0A1J1HI61 A0A2H2IN21 A0A2P6L3P9 A0A0V1MZF6 A0A0V1J3G8 A0A0V1H1U7 A0A087TD78 A0A0V1CXK4 A0A2W1B8Z9 A0A016VC00 E5S2I0 A0A016VAF8 A0A016V9B6 A0A368GXV9 A0A0D6LGI9 A0A090N0V2 A0A1Y1KW09 A0A2P6LAR8 A0A0V1NT84 A0A0V0Z760 A0A0V1LSL0 A0A0V0VIZ3 A0A0V0U5N5 A0A1I8A2H2 A0A0V1B9Z3 A0A0V1B9Y2 A0A0L7LIN3 A0A0V0WZ71 A0A0V1FGY8 A0A0V1J3E7 A0A131ZVD5 A0A194Q2H2 A0A0V0WZ93 A0A368F3L2 A0A0V0Z7C1 A0A0V0Z7M5 A0A087UJV8 A0A1I7Z116 A0A090L0U6 A0A0R3UUA3 A0A1I8A6G1 A0A0V0Z791 A0A0N4Y782 A0A0V0Z793 A0A0N5A6N4 A0A0V0U5E1 A0A194Q8M7 A0A2A4JCJ6 A0A336LED8 A0A0V1EPP9 A0A0K0EAF5 A0A0V0S515 A0A0V1EPP1 H2L2K9 A0A212EIN9 A0A0V1EPN2 A0A2A4J7B9 E4WQ01 D2A397 A0A1I7T4K9 A0A2L2YKC7 H9JHU8 A0A260Z7K7 B0XBB5 H3G2Y3 A0A0V0S563 E4XIX0 A0A2P6L3Q8 A0A0C2D8P6 A0A194QS46 A0A0V0S5K9 A0A1I8BL05 A0A016VB53 A0A2L2XYH3 A0A2L2XYN6 A0A0V0U579 A0A194QS41 C3Y5D2 A0A0C2CJT7 B7P7B7
Pubmed
EMBL
GG666636
EEN46977.1
CVRI01000021
CRK91692.1
UZAM01013500
VDP28241.1
+ More
ODYU01007799 SOQ50866.1 CVRI01000059 CRL03442.1 KN729227 KIH62569.1 JI180803 ADY48529.1 ODYU01002951 SOQ41102.1 CVRI01000004 CRK87701.1 MWRG01001996 PRD33208.1 JYDO01000022 KRZ77159.1 JYDS01000045 KRZ29499.1 JYDP01000166 KRZ04345.1 KK114672 KFM63067.1 JYDI01000077 KRY53944.1 KZ150221 PZC72102.1 JARK01001350 EYC24263.1 EYC24261.1 EYC24264.1 JOJR01000037 RCN49202.1 KE125129 EPB71200.1 LN609530 CEF71333.1 GEZM01072176 JAV65593.1 MWRG01000603 PRD35666.1 JYDM01000104 KRZ87210.1 JYDQ01000343 KRY08320.1 JYDW01000009 KRZ62443.1 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 JYDH01000076 KRY33812.1 KRY33814.1 JTDY01000922 KOB75423.1 JYDK01000035 KRX81073.1 JYDT01000094 KRY85257.1 KRZ29498.1 JXLN01002281 KPM02581.1 KQ459562 KPI99762.1 KRX81072.1 JOJR01011584 RCN25505.1 KRY08317.1 KRY08323.1 KK120157 KFM77647.1 LN609496 CEF61712.1 KRY08319.1 UYSL01020651 VDL75592.1 KRY08321.1 KRX46473.1 KPI99760.1 NWSH01001919 PCG69681.1 UFQS01003067 UFQT01003067 SSX15127.1 SSX34506.1 JYDR01000016 KRY75718.1 JYDL01000037 KRX21785.1 KRY75722.1 BX284605 CCE71937.1 AGBW02014600 OWR41356.1 KRY75723.1 NWSH01002542 PCG68017.1 FN653015 CBY20035.1 KQ971338 EFA02281.1 IAAA01030941 LAA08563.1 BABH01029524 NMWX01000238 OZF81607.1 DS232623 EDS44206.1 KRX21783.1 FN653056 FN655682 CBY24654.1 CBY39860.1 PRD33209.1 KN727256 KIH66103.1 KQ461175 KPJ07805.1 KRX21786.1 EYC24262.1 IAAA01002395 IAAA01002396 LAA01006.1 IAAA01002394 LAA00998.1 KRX46474.1 KPJ07800.1 GG666487 EEN64179.1 KN751001 KIH50047.1 ABJB010733936 ABJB010950942 DS651071 EEC02489.1
ODYU01007799 SOQ50866.1 CVRI01000059 CRL03442.1 KN729227 KIH62569.1 JI180803 ADY48529.1 ODYU01002951 SOQ41102.1 CVRI01000004 CRK87701.1 MWRG01001996 PRD33208.1 JYDO01000022 KRZ77159.1 JYDS01000045 KRZ29499.1 JYDP01000166 KRZ04345.1 KK114672 KFM63067.1 JYDI01000077 KRY53944.1 KZ150221 PZC72102.1 JARK01001350 EYC24263.1 EYC24261.1 EYC24264.1 JOJR01000037 RCN49202.1 KE125129 EPB71200.1 LN609530 CEF71333.1 GEZM01072176 JAV65593.1 MWRG01000603 PRD35666.1 JYDM01000104 KRZ87210.1 JYDQ01000343 KRY08320.1 JYDW01000009 KRZ62443.1 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 JYDH01000076 KRY33812.1 KRY33814.1 JTDY01000922 KOB75423.1 JYDK01000035 KRX81073.1 JYDT01000094 KRY85257.1 KRZ29498.1 JXLN01002281 KPM02581.1 KQ459562 KPI99762.1 KRX81072.1 JOJR01011584 RCN25505.1 KRY08317.1 KRY08323.1 KK120157 KFM77647.1 LN609496 CEF61712.1 KRY08319.1 UYSL01020651 VDL75592.1 KRY08321.1 KRX46473.1 KPI99760.1 NWSH01001919 PCG69681.1 UFQS01003067 UFQT01003067 SSX15127.1 SSX34506.1 JYDR01000016 KRY75718.1 JYDL01000037 KRX21785.1 KRY75722.1 BX284605 CCE71937.1 AGBW02014600 OWR41356.1 KRY75723.1 NWSH01002542 PCG68017.1 FN653015 CBY20035.1 KQ971338 EFA02281.1 IAAA01030941 LAA08563.1 BABH01029524 NMWX01000238 OZF81607.1 DS232623 EDS44206.1 KRX21783.1 FN653056 FN655682 CBY24654.1 CBY39860.1 PRD33209.1 KN727256 KIH66103.1 KQ461175 KPJ07805.1 KRX21786.1 EYC24262.1 IAAA01002395 IAAA01002396 LAA01006.1 IAAA01002394 LAA00998.1 KRX46474.1 KPJ07800.1 GG666487 EEN64179.1 KN751001 KIH50047.1 ABJB010733936 ABJB010950942 DS651071 EEC02489.1
Proteomes
UP000001554
UP000183832
UP000050793
UP000005237
UP000054843
UP000054805
+ More
UP000055024 UP000054359 UP000054653 UP000024635 UP000252519 UP000035682 UP000054924 UP000054783 UP000054721 UP000054681 UP000055048 UP000095287 UP000054776 UP000037510 UP000054673 UP000054995 UP000053268 UP000038043 UP000271162 UP000038045 UP000218220 UP000054632 UP000035681 UP000054630 UP000001940 UP000007151 UP000007266 UP000095282 UP000005204 UP000216624 UP000002320 UP000005239 UP000053240 UP000095281 UP000001555
UP000055024 UP000054359 UP000054653 UP000024635 UP000252519 UP000035682 UP000054924 UP000054783 UP000054721 UP000054681 UP000055048 UP000095287 UP000054776 UP000037510 UP000054673 UP000054995 UP000053268 UP000038043 UP000271162 UP000038045 UP000218220 UP000054632 UP000035681 UP000054630 UP000001940 UP000007151 UP000007266 UP000095282 UP000005204 UP000216624 UP000002320 UP000005239 UP000053240 UP000095281 UP000001555
PRIDE
Pfam
Interpro
IPR036084
Ser_inhib-like_sf
+ More
IPR002919 TIL_dom
IPR000742 EGF-like_dom
IPR000772 Ricin_B_lectin
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR016092 FeS_cluster_insertion
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR035903 HesB-like_dom_sf
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR004855 TFIIA_asu/bsu
IPR004843 Calcineurin-like_PHP_ApaH
IPR013026 TPR-contain_dom
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR000361 FeS_biogenesis
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR002223 Kunitz_BPTI
IPR003645 Fol_N
IPR000922 Lectin_gal-bd_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR016187 CTDL_fold
IPR001846 VWF_type-D
IPR025615 TILa_dom
IPR035234 IgGFc-bd_N
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR016186 C-type_lectin-like/link_sf
IPR014853 Unchr_dom_Cys-rich
IPR009030 Growth_fac_rcpt_cys_sf
IPR024731 EGF_dom
IPR001881 EGF-like_Ca-bd_dom
IPR001304 C-type_lectin-like
IPR002919 TIL_dom
IPR000742 EGF-like_dom
IPR000772 Ricin_B_lectin
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR016092 FeS_cluster_insertion
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR035903 HesB-like_dom_sf
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR004855 TFIIA_asu/bsu
IPR004843 Calcineurin-like_PHP_ApaH
IPR013026 TPR-contain_dom
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR000361 FeS_biogenesis
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR002223 Kunitz_BPTI
IPR003645 Fol_N
IPR000922 Lectin_gal-bd_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR016187 CTDL_fold
IPR001846 VWF_type-D
IPR025615 TILa_dom
IPR035234 IgGFc-bd_N
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR016186 C-type_lectin-like/link_sf
IPR014853 Unchr_dom_Cys-rich
IPR009030 Growth_fac_rcpt_cys_sf
IPR024731 EGF_dom
IPR001881 EGF-like_Ca-bd_dom
IPR001304 C-type_lectin-like
SUPFAM
ProteinModelPortal
C3ZK78
A0A1J1HWF2
A0A183J2C6
A0A2H1WD28
A0A1J1IUR1
A0A0C2GUB0
+ More
F1LEH5 A0A2H1VKZ4 A0A1J1HI61 A0A2H2IN21 A0A2P6L3P9 A0A0V1MZF6 A0A0V1J3G8 A0A0V1H1U7 A0A087TD78 A0A0V1CXK4 A0A2W1B8Z9 A0A016VC00 E5S2I0 A0A016VAF8 A0A016V9B6 A0A368GXV9 A0A0D6LGI9 A0A090N0V2 A0A1Y1KW09 A0A2P6LAR8 A0A0V1NT84 A0A0V0Z760 A0A0V1LSL0 A0A0V0VIZ3 A0A0V0U5N5 A0A1I8A2H2 A0A0V1B9Z3 A0A0V1B9Y2 A0A0L7LIN3 A0A0V0WZ71 A0A0V1FGY8 A0A0V1J3E7 A0A131ZVD5 A0A194Q2H2 A0A0V0WZ93 A0A368F3L2 A0A0V0Z7C1 A0A0V0Z7M5 A0A087UJV8 A0A1I7Z116 A0A090L0U6 A0A0R3UUA3 A0A1I8A6G1 A0A0V0Z791 A0A0N4Y782 A0A0V0Z793 A0A0N5A6N4 A0A0V0U5E1 A0A194Q8M7 A0A2A4JCJ6 A0A336LED8 A0A0V1EPP9 A0A0K0EAF5 A0A0V0S515 A0A0V1EPP1 H2L2K9 A0A212EIN9 A0A0V1EPN2 A0A2A4J7B9 E4WQ01 D2A397 A0A1I7T4K9 A0A2L2YKC7 H9JHU8 A0A260Z7K7 B0XBB5 H3G2Y3 A0A0V0S563 E4XIX0 A0A2P6L3Q8 A0A0C2D8P6 A0A194QS46 A0A0V0S5K9 A0A1I8BL05 A0A016VB53 A0A2L2XYH3 A0A2L2XYN6 A0A0V0U579 A0A194QS41 C3Y5D2 A0A0C2CJT7 B7P7B7
F1LEH5 A0A2H1VKZ4 A0A1J1HI61 A0A2H2IN21 A0A2P6L3P9 A0A0V1MZF6 A0A0V1J3G8 A0A0V1H1U7 A0A087TD78 A0A0V1CXK4 A0A2W1B8Z9 A0A016VC00 E5S2I0 A0A016VAF8 A0A016V9B6 A0A368GXV9 A0A0D6LGI9 A0A090N0V2 A0A1Y1KW09 A0A2P6LAR8 A0A0V1NT84 A0A0V0Z760 A0A0V1LSL0 A0A0V0VIZ3 A0A0V0U5N5 A0A1I8A2H2 A0A0V1B9Z3 A0A0V1B9Y2 A0A0L7LIN3 A0A0V0WZ71 A0A0V1FGY8 A0A0V1J3E7 A0A131ZVD5 A0A194Q2H2 A0A0V0WZ93 A0A368F3L2 A0A0V0Z7C1 A0A0V0Z7M5 A0A087UJV8 A0A1I7Z116 A0A090L0U6 A0A0R3UUA3 A0A1I8A6G1 A0A0V0Z791 A0A0N4Y782 A0A0V0Z793 A0A0N5A6N4 A0A0V0U5E1 A0A194Q8M7 A0A2A4JCJ6 A0A336LED8 A0A0V1EPP9 A0A0K0EAF5 A0A0V0S515 A0A0V1EPP1 H2L2K9 A0A212EIN9 A0A0V1EPN2 A0A2A4J7B9 E4WQ01 D2A397 A0A1I7T4K9 A0A2L2YKC7 H9JHU8 A0A260Z7K7 B0XBB5 H3G2Y3 A0A0V0S563 E4XIX0 A0A2P6L3Q8 A0A0C2D8P6 A0A194QS46 A0A0V0S5K9 A0A1I8BL05 A0A016VB53 A0A2L2XYH3 A0A2L2XYN6 A0A0V0U579 A0A194QS41 C3Y5D2 A0A0C2CJT7 B7P7B7
Ontologies
GO
PANTHER
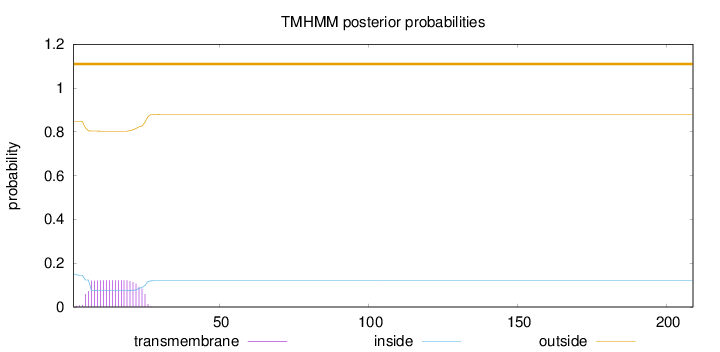
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.996973
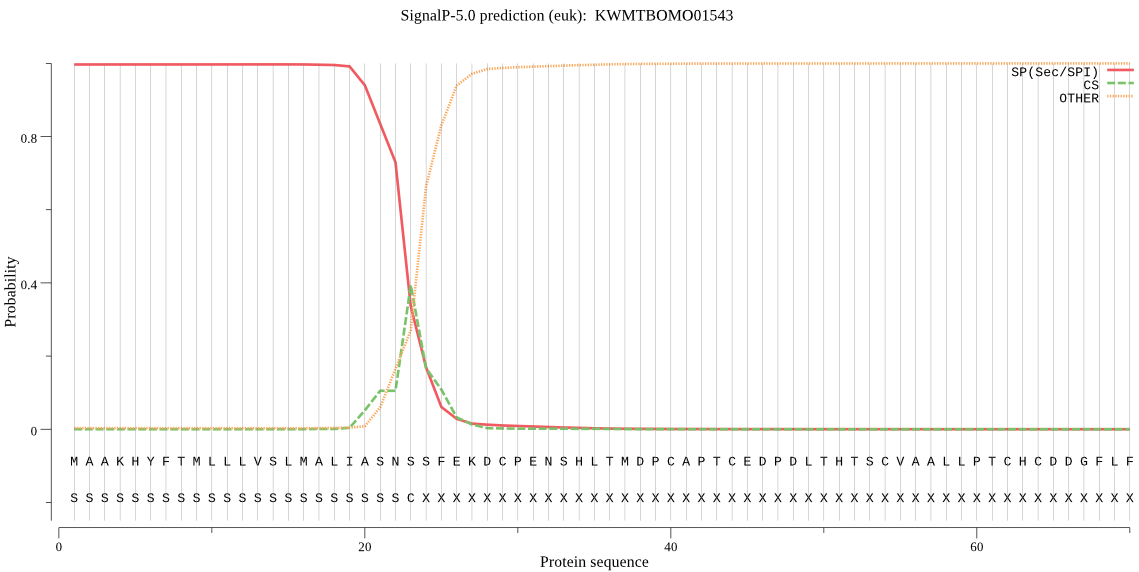
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.31861
Exp number, first 60 AAs:
2.31861
Total prob of N-in:
0.15236
outside
1 - 209
Population Genetic Test Statistics
Pi
278.617538
Theta
189.159126
Tajima's D
1.513629
CLR
122.620415
CSRT
0.793860306984651
Interpretation
Uncertain
