Gene
KWMTBOMO01542
Pre Gene Modal
BGIBMGA009094
Annotation
fungal_protease_inhibitor_[Bombyx_mori]
Full name
Fungal protease inhibitor F
Location in the cell
PlasmaMembrane Reliability : 1.291
Sequence
CDS
ATGGCCGCCAAACAATACTTTATCGTGTTCCTGATTGTCGCCGTGATGGCTCTCGAGGCCAGCACTACCGAATATGGATGCCCTGAAAATGCCCATTGGACTGATGACCCCTGCGTGAGGACCTGCGACGACCCGTACCTGACTAACACCGCTTGCGTCGGCGCCCTCATTCAAACCTGCCACTGTAACGACGGCTTAGTCTTCAACGCGGACAGAAAATGTGTGCCCATTTCTGATTGTTAA
Protein
MAAKQYFIVFLIVAVMALEASTTEYGCPENAHWTDDPCVRTCDDPYLTNTACVGALIQTCHCNDGLVFNADRKCVPISDC
Summary
Similarity
Belongs to the protease inhibitor I40 family.
Keywords
Complete proteome
Direct protein sequencing
Disulfide bond
Protease inhibitor
Reference proteome
Secreted
Serine protease inhibitor
Signal
Feature
chain Fungal protease inhibitor F
Uniprot
Pubmed
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Ontologies
Topology
Subcellular location
Secreted
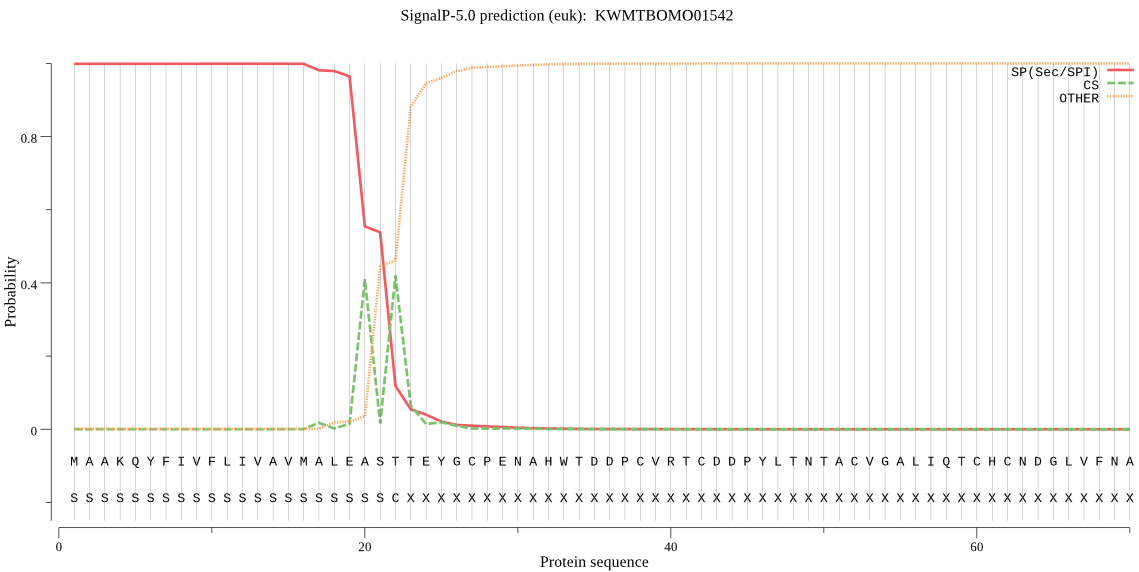
SignalP
Position: 1 - 22,
Likelihood: 0.999008
Length:
80
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.39032
Exp number, first 60 AAs:
7.3502
Total prob of N-in:
0.51161
outside
1 - 80

Population Genetic Test Statistics
Pi
173.356503
Theta
193.947234
Tajima's D
-0.585498
CLR
0.002437
CSRT
0.216189190540473
Interpretation
Uncertain
