Gene
KWMTBOMO01538
Pre Gene Modal
BGIBMGA009072
Annotation
fungal_protease_inhibitor_[Bombyx_mori]
Full name
Fungal protease inhibitor F
+ More
IgGFc-binding protein
IgGFc-binding protein
Alternative Name
Fcgamma-binding protein antigen
Location in the cell
Mitochondrial Reliability : 1.256 Nuclear Reliability : 1.052
Sequence
CDS
ATGGCCGCCAAACAATACTTTATCGTGTTCCTGATTGTCGCCCTGATGACTCTTGGTGCCAGCACTGCTGACGATGGGTGTCCCGAGAATGCTCATTGGACTGATGACCCCTGCGCGAAGACCTGCGACGACCCATACTTGACTAACACCGTTTGCATCGCCGCCCTCATTCCAACCTGCCACTGTAACAGCGGCTTAGTCTTCAACGCGGAGAGAAAATGTGTACCCATTTCGGATTGTTAA
Protein
MAAKQYFIVFLIVALMTLGASTADDGCPENAHWTDDPCAKTCDDPYLTNTVCIAALIPTCHCNSGLVFNAERKCVPISDC
Summary
Description
May be involved in the maintenance of the mucosal structure as a gel-like component of the mucosa.
Subunit
Interacts with the Fc portion of IgG and with MUC2.
Similarity
Belongs to the protease inhibitor I40 family.
Keywords
Complete proteome
Direct protein sequencing
Disulfide bond
Protease inhibitor
Reference proteome
Secreted
Serine protease inhibitor
Signal
Glycoprotein
Polymorphism
Repeat
Feature
chain Fungal protease inhibitor F
sequence variant In dbSNP:rs34181317.
sequence variant In dbSNP:rs34181317.
Uniprot
Pubmed
EMBL
BABH01029521
BABH01029516
KP987266
AKJ54534.1
BABH01029514
KP987267
+ More
AKJ54535.1 BABH01029517 KC153969 AGE81791.1 BABH01029512 S83181 D38075 PCDA01000019 MAA51344.1 GAKP01009193 JAC49759.1 JRES01000712 KNC29045.1 NHHK01000027 OUW20087.1 ADTU01005853 NIPN01000007 OZG21929.1 QQZZ01000181 RMZ36946.1 EQ963482 EED47799.1 KQ976738 KYM75719.1 JZDT01000492 KOC13483.1 KK115573 KFM65461.1 QOIP01000009 RLU18198.1 DS268420 LFJK02001379 EFO87795.1 POM31849.1 NMWX01000009 OZF94174.1 GL379809 EGT42507.1 D84239 AC006950 AC007842 AC011536 PDUG01000005 PIC22215.1
AKJ54535.1 BABH01029517 KC153969 AGE81791.1 BABH01029512 S83181 D38075 PCDA01000019 MAA51344.1 GAKP01009193 JAC49759.1 JRES01000712 KNC29045.1 NHHK01000027 OUW20087.1 ADTU01005853 NIPN01000007 OZG21929.1 QQZZ01000181 RMZ36946.1 EQ963482 EED47799.1 KQ976738 KYM75719.1 JZDT01000492 KOC13483.1 KK115573 KFM65461.1 QOIP01000009 RLU18198.1 DS268420 LFJK02001379 EFO87795.1 POM31849.1 NMWX01000009 OZF94174.1 GL379809 EGT42507.1 D84239 AC006950 AC007842 AC011536 PDUG01000005 PIC22215.1
Proteomes
Pfam
Interpro
IPR002919
TIL_dom
+ More
IPR036084 Ser_inhib-like_sf
IPR032675 LRR_dom_sf
IPR016134 Dockerin_dom
IPR036439 Dockerin_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR003366 CUB-like_dom
IPR001888 Transposase_1
IPR036388 WH-like_DNA-bd_sf
IPR014853 Unchr_dom_Cys-rich
IPR001846 VWF_type-D
IPR035234 IgGFc-bd_N
IPR000742 EGF-like_dom
IPR001007 VWF_dom
IPR025615 TILa_dom
IPR003645 Fol_N
IPR036084 Ser_inhib-like_sf
IPR032675 LRR_dom_sf
IPR016134 Dockerin_dom
IPR036439 Dockerin_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR003366 CUB-like_dom
IPR001888 Transposase_1
IPR036388 WH-like_DNA-bd_sf
IPR014853 Unchr_dom_Cys-rich
IPR001846 VWF_type-D
IPR035234 IgGFc-bd_N
IPR000742 EGF-like_dom
IPR001007 VWF_dom
IPR025615 TILa_dom
IPR003645 Fol_N
Gene 3D
ProteinModelPortal
Ontologies
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 23,
Likelihood: 0.999289
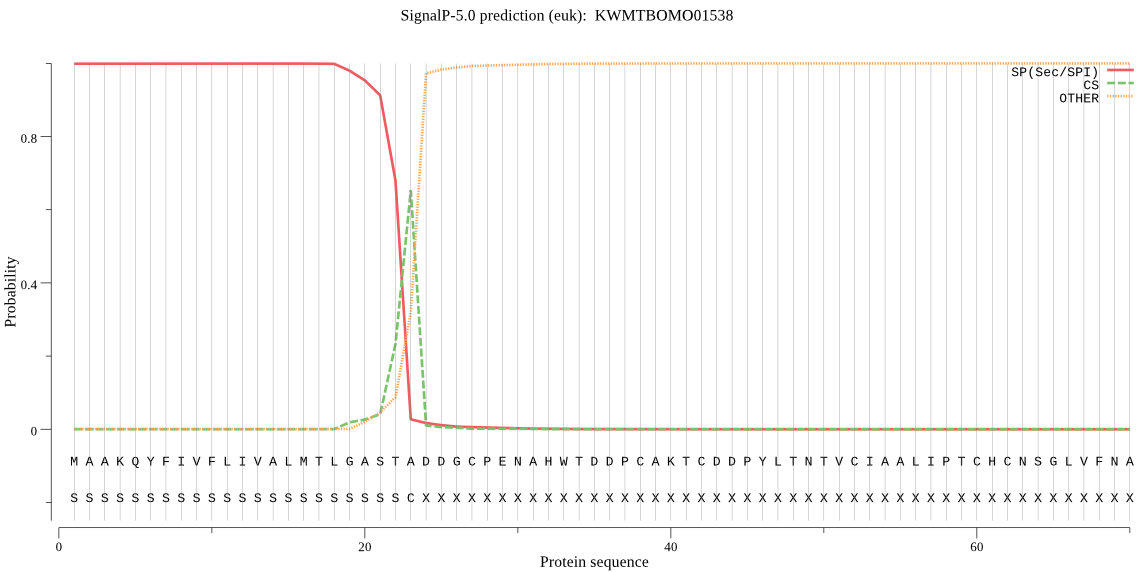
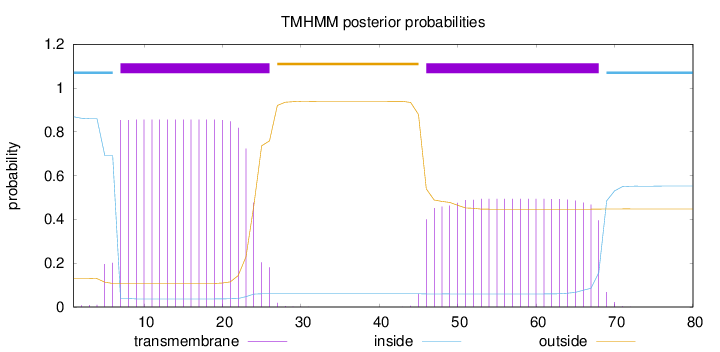
Length:
80
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
26.80131
Exp number, first 60 AAs:
22.91252
Total prob of N-in:
0.86898
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 45
TMhelix
46 - 68
inside
69 - 80
Population Genetic Test Statistics
Pi
146.139951
Theta
153.846756
Tajima's D
-0.157493
CLR
0.200013
CSRT
0.323533823308835
Interpretation
Uncertain
