Gene
KWMTBOMO01536 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009096
Annotation
PREDICTED:_inducible_metalloproteinase_inhibitor_protein-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.666
Sequence
CDS
ATGTGCATTACCGGCTGCTTCTGCGAGGAGGGACAAGTTAACAACGACAACGGGGTCTGCGTGAACTTAGCCGACTGCCCTCAAGCCGCTTCCGCATACAAGCTGCAAACAACAGAACCACGATTCGATGGAGGAAAGTGCCCTCAAAACGAGGAGTACAAATTCTGCGAGCCCTGTAACAGGACTTGCGAGAATCCGTTCCCGGTGTGTCCAGCGCAGTGCGCCCGAGGATGTTTCTGTAAAGACGGTCTGGTGAGGGACAAAGACGGAAAATGCGTGGAATTGGAACAGTGCTCAAACTTAAAACATCTTAAGCTTGGAGAAAACTACAAACAGCCAATCCCTTCCCGCGTCAACTGCGGTCCCTATCAAGTGTACAAGAAGTGTGGCACCTGTGACAAAACCTGCAGTAATCCTAACCCTGTGTGCAACCAAGCCTGCCAGAGAGGATGCTTCTGCCAAGAGGGCTATGTTAAATCTGTTCACGGAAGCTGCGTGAAACAGGAAGATTGTCCTGATGGTTCGTAG
Protein
MCITGCFCEEGQVNNDNGVCVNLADCPQAASAYKLQTTEPRFDGGKCPQNEEYKFCEPCNRTCENPFPVCPAQCARGCFCKDGLVRDKDGKCVELEQCSNLKHLKLGENYKQPIPSRVNCGPYQVYKKCGTCDKTCSNPNPVCNQACQRGCFCQEGYVKSVHGSCVKQEDCPDGS
Summary
Uniprot
H9JHU8
A0A2H1WD28
A0A2A4JCJ6
A0A0L7LIN3
A0A194Q2H2
A0A194QS46
+ More
A0A0L7K498 A0A087UJV8 A0A0V1EPP9 A0A0V0Z7C1 A0A0V0S515 A0A2L2XYH3 A0A2L2XYN6 A0A182NIS5 A0A087TD78 B0XBB5 A0A1I7Z116 A0A2P6L3Q8 A0A1I7Z0X9 A0A0V0Z7M5 A0A1I7Z1E9 A0A0V1FGY8 A0A0V0Z791 A0A0V0S563 A0A0V0S5K9 T1L4R5 A0A2A4JLM8 A0A2P6KNP2 A0A0V1CXK4 A0A2Z6FL32 A0A194Q8M7 A0A194QS41 E5S2I0 A0A2P6L3P9 A0A0V0Z760 A0A087UDS0 A0A183J2C6 A0A0V0Z793 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1B9Y2 A0A0V1B9Z3 A0A0V1NT84 A0A0V0U5E1 A0A0V0WZ71 A0A0V1LSL0 A0A2H1WS62 A0A0V1H1U7 H9JHS7 A0A0V0U579 A0A0V0WZ93 A0A1I7ZRE6 A0A2P6L3P8 A0A0V1MZF6 A0A0V1EPP1 A0A0V1J3G8 A0A0V1J3E7 A0A0V1EPN2 E5T4Z6 A0A0N4X8U1 A0A1I8A6G1 A0A1Y3EJT5 A0A016VB53 A0A1I7YSE4 A0A0N5BI72 A0A2A2JIB4 A0A1I8A2H2 A0A0N5BYG0 A0A0M3J546 A0A0C2GUB0 A0A2P6LAR8 E7D188 A0A0D6LGI9 A0A016VAF8 A0A2P6L3T3 A0A2H1VNG5 A0A183UX28 D2A397 A0A2L2XZL2 A0A0L7LIA1 A0A0M3J1Y0 A0A2A6C9V2 A0A3P7S1D0 A0A0N4TEC4 A0A1I7ZNL3 A0A1I7ZPH5 A0A3P7E4K6 A0A368GXV9 A0A0K0FEA5 A0A0K0JE24 A0A0B2V3K6 A0A131YYX3 A0A2L2Y735 A0A182RDG1 B7P7B7 A0A147BD52 A0A0N4Y782 A0A087TK17 A0A2L2Y4K9
A0A0L7K498 A0A087UJV8 A0A0V1EPP9 A0A0V0Z7C1 A0A0V0S515 A0A2L2XYH3 A0A2L2XYN6 A0A182NIS5 A0A087TD78 B0XBB5 A0A1I7Z116 A0A2P6L3Q8 A0A1I7Z0X9 A0A0V0Z7M5 A0A1I7Z1E9 A0A0V1FGY8 A0A0V0Z791 A0A0V0S563 A0A0V0S5K9 T1L4R5 A0A2A4JLM8 A0A2P6KNP2 A0A0V1CXK4 A0A2Z6FL32 A0A194Q8M7 A0A194QS41 E5S2I0 A0A2P6L3P9 A0A0V0Z760 A0A087UDS0 A0A183J2C6 A0A0V0Z793 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1B9Y2 A0A0V1B9Z3 A0A0V1NT84 A0A0V0U5E1 A0A0V0WZ71 A0A0V1LSL0 A0A2H1WS62 A0A0V1H1U7 H9JHS7 A0A0V0U579 A0A0V0WZ93 A0A1I7ZRE6 A0A2P6L3P8 A0A0V1MZF6 A0A0V1EPP1 A0A0V1J3G8 A0A0V1J3E7 A0A0V1EPN2 E5T4Z6 A0A0N4X8U1 A0A1I8A6G1 A0A1Y3EJT5 A0A016VB53 A0A1I7YSE4 A0A0N5BI72 A0A2A2JIB4 A0A1I8A2H2 A0A0N5BYG0 A0A0M3J546 A0A0C2GUB0 A0A2P6LAR8 E7D188 A0A0D6LGI9 A0A016VAF8 A0A2P6L3T3 A0A2H1VNG5 A0A183UX28 D2A397 A0A2L2XZL2 A0A0L7LIA1 A0A0M3J1Y0 A0A2A6C9V2 A0A3P7S1D0 A0A0N4TEC4 A0A1I7ZNL3 A0A1I7ZPH5 A0A3P7E4K6 A0A368GXV9 A0A0K0FEA5 A0A0K0JE24 A0A0B2V3K6 A0A131YYX3 A0A2L2Y735 A0A182RDG1 B7P7B7 A0A147BD52 A0A0N4Y782 A0A087TK17 A0A2L2Y4K9
Pubmed
EMBL
BABH01029524
ODYU01007799
SOQ50866.1
NWSH01001919
PCG69681.1
JTDY01000922
+ More
KOB75423.1 KQ459562 KPI99762.1 KQ461175 KPJ07805.1 JTDY01011380 KOB56694.1 KK120157 KFM77647.1 JYDR01000016 KRY75718.1 JYDQ01000343 KRY08317.1 JYDL01000037 KRX21785.1 IAAA01002395 IAAA01002396 LAA01006.1 IAAA01002394 LAA00998.1 KK114672 KFM63067.1 DS232623 EDS44206.1 MWRG01001996 PRD33209.1 KRY08323.1 JYDT01000094 KRY85257.1 KRY08319.1 KRX21783.1 KRX21786.1 CAEY01001161 NWSH01001153 PCG72343.1 MWRG01008016 PRD27941.1 JYDI01000077 KRY53944.1 LC384847 BBE28990.1 KPI99760.1 KPJ07800.1 PRD33208.1 KRY08320.1 KK119375 KFM75509.1 UZAM01013500 VDP28241.1 KRY08321.1 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 JYDH01000076 KRY33814.1 KRY33812.1 JYDM01000104 KRZ87210.1 KRX46473.1 JYDK01000035 KRX81073.1 JYDW01000009 KRZ62443.1 ODYU01010657 SOQ55910.1 JYDP01000166 KRZ04345.1 BABH01029478 KRX46474.1 KRX81072.1 PRD33210.1 JYDO01000022 KRZ77159.1 KRY75722.1 JYDS01000045 KRZ29499.1 KRZ29498.1 KRY75723.1 UZAF01022568 VDO85928.1 LVZM01011071 OUC44980.1 JARK01001350 EYC24262.1 LIAE01010412 PAV61486.1 UYRR01003431 VDK20089.1 KN729227 KIH62569.1 MWRG01000603 PRD35666.1 HQ005836 ADV40132.1 KE125129 EPB71200.1 EYC24261.1 PRD33207.1 ODYU01003504 SOQ42379.1 UYWY01021537 VDM44369.1 KQ971338 EFA02281.1 IAAA01000101 LAA01399.1 JTDY01001071 KOB74966.1 UYRR01001489 VDK18738.1 ABKE03000053 PDM74992.1 UZAD01006078 VDN87712.1 UYWW01001863 VDM11094.1 JOJR01000037 RCN49202.1 JPKZ01002570 KHN76064.1 GEDV01004360 JAP84197.1 IAAA01007013 LAA03120.1 ABJB010733936 ABJB010950942 DS651071 EEC02489.1 GEGO01006701 JAR88703.1 UYSL01020651 VDL75592.1 KK115573 KFM65456.1 IAAA01007012 LAA03116.1
KOB75423.1 KQ459562 KPI99762.1 KQ461175 KPJ07805.1 JTDY01011380 KOB56694.1 KK120157 KFM77647.1 JYDR01000016 KRY75718.1 JYDQ01000343 KRY08317.1 JYDL01000037 KRX21785.1 IAAA01002395 IAAA01002396 LAA01006.1 IAAA01002394 LAA00998.1 KK114672 KFM63067.1 DS232623 EDS44206.1 MWRG01001996 PRD33209.1 KRY08323.1 JYDT01000094 KRY85257.1 KRY08319.1 KRX21783.1 KRX21786.1 CAEY01001161 NWSH01001153 PCG72343.1 MWRG01008016 PRD27941.1 JYDI01000077 KRY53944.1 LC384847 BBE28990.1 KPI99760.1 KPJ07800.1 PRD33208.1 KRY08320.1 KK119375 KFM75509.1 UZAM01013500 VDP28241.1 KRY08321.1 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 JYDH01000076 KRY33814.1 KRY33812.1 JYDM01000104 KRZ87210.1 KRX46473.1 JYDK01000035 KRX81073.1 JYDW01000009 KRZ62443.1 ODYU01010657 SOQ55910.1 JYDP01000166 KRZ04345.1 BABH01029478 KRX46474.1 KRX81072.1 PRD33210.1 JYDO01000022 KRZ77159.1 KRY75722.1 JYDS01000045 KRZ29499.1 KRZ29498.1 KRY75723.1 UZAF01022568 VDO85928.1 LVZM01011071 OUC44980.1 JARK01001350 EYC24262.1 LIAE01010412 PAV61486.1 UYRR01003431 VDK20089.1 KN729227 KIH62569.1 MWRG01000603 PRD35666.1 HQ005836 ADV40132.1 KE125129 EPB71200.1 EYC24261.1 PRD33207.1 ODYU01003504 SOQ42379.1 UYWY01021537 VDM44369.1 KQ971338 EFA02281.1 IAAA01000101 LAA01399.1 JTDY01001071 KOB74966.1 UYRR01001489 VDK18738.1 ABKE03000053 PDM74992.1 UZAD01006078 VDN87712.1 UYWW01001863 VDM11094.1 JOJR01000037 RCN49202.1 JPKZ01002570 KHN76064.1 GEDV01004360 JAP84197.1 IAAA01007013 LAA03120.1 ABJB010733936 ABJB010950942 DS651071 EEC02489.1 GEGO01006701 JAR88703.1 UYSL01020651 VDL75592.1 KK115573 KFM65456.1 IAAA01007012 LAA03116.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000054359
+ More
UP000054632 UP000054783 UP000054630 UP000075884 UP000002320 UP000095287 UP000054995 UP000015104 UP000054653 UP000050793 UP000054681 UP000055048 UP000054776 UP000054924 UP000054673 UP000054721 UP000055024 UP000054843 UP000054805 UP000038042 UP000268014 UP000243006 UP000024635 UP000046392 UP000218231 UP000036680 UP000050794 UP000267007 UP000007266 UP000232936 UP000278627 UP000038020 UP000270924 UP000252519 UP000035680 UP000006672 UP000031036 UP000075900 UP000001555 UP000038043 UP000271162
UP000054632 UP000054783 UP000054630 UP000075884 UP000002320 UP000095287 UP000054995 UP000015104 UP000054653 UP000050793 UP000054681 UP000055048 UP000054776 UP000054924 UP000054673 UP000054721 UP000055024 UP000054843 UP000054805 UP000038042 UP000268014 UP000243006 UP000024635 UP000046392 UP000218231 UP000036680 UP000050794 UP000267007 UP000007266 UP000232936 UP000278627 UP000038020 UP000270924 UP000252519 UP000035680 UP000006672 UP000031036 UP000075900 UP000001555 UP000038043 UP000271162
Pfam
Interpro
IPR036084
Ser_inhib-like_sf
+ More
IPR002919 TIL_dom
IPR000742 EGF-like_dom
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR016092 FeS_cluster_insertion
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR035903 HesB-like_dom_sf
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR004855 TFIIA_asu/bsu
IPR004843 Calcineurin-like_PHP_ApaH
IPR013026 TPR-contain_dom
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR000361 FeS_biogenesis
IPR020901 Prtase_inh_Kunz-CS
IPR036880 Kunitz_BPTI_sf
IPR002223 Kunitz_BPTI
IPR003645 Fol_N
IPR031305 Casein_CS
IPR002919 TIL_dom
IPR000742 EGF-like_dom
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR016092 FeS_cluster_insertion
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR035903 HesB-like_dom_sf
IPR019734 TPR_repeat
IPR011990 TPR-like_helical_dom_sf
IPR004855 TFIIA_asu/bsu
IPR004843 Calcineurin-like_PHP_ApaH
IPR013026 TPR-contain_dom
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR000361 FeS_biogenesis
IPR020901 Prtase_inh_Kunz-CS
IPR036880 Kunitz_BPTI_sf
IPR002223 Kunitz_BPTI
IPR003645 Fol_N
IPR031305 Casein_CS
Gene 3D
ProteinModelPortal
H9JHU8
A0A2H1WD28
A0A2A4JCJ6
A0A0L7LIN3
A0A194Q2H2
A0A194QS46
+ More
A0A0L7K498 A0A087UJV8 A0A0V1EPP9 A0A0V0Z7C1 A0A0V0S515 A0A2L2XYH3 A0A2L2XYN6 A0A182NIS5 A0A087TD78 B0XBB5 A0A1I7Z116 A0A2P6L3Q8 A0A1I7Z0X9 A0A0V0Z7M5 A0A1I7Z1E9 A0A0V1FGY8 A0A0V0Z791 A0A0V0S563 A0A0V0S5K9 T1L4R5 A0A2A4JLM8 A0A2P6KNP2 A0A0V1CXK4 A0A2Z6FL32 A0A194Q8M7 A0A194QS41 E5S2I0 A0A2P6L3P9 A0A0V0Z760 A0A087UDS0 A0A183J2C6 A0A0V0Z793 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1B9Y2 A0A0V1B9Z3 A0A0V1NT84 A0A0V0U5E1 A0A0V0WZ71 A0A0V1LSL0 A0A2H1WS62 A0A0V1H1U7 H9JHS7 A0A0V0U579 A0A0V0WZ93 A0A1I7ZRE6 A0A2P6L3P8 A0A0V1MZF6 A0A0V1EPP1 A0A0V1J3G8 A0A0V1J3E7 A0A0V1EPN2 E5T4Z6 A0A0N4X8U1 A0A1I8A6G1 A0A1Y3EJT5 A0A016VB53 A0A1I7YSE4 A0A0N5BI72 A0A2A2JIB4 A0A1I8A2H2 A0A0N5BYG0 A0A0M3J546 A0A0C2GUB0 A0A2P6LAR8 E7D188 A0A0D6LGI9 A0A016VAF8 A0A2P6L3T3 A0A2H1VNG5 A0A183UX28 D2A397 A0A2L2XZL2 A0A0L7LIA1 A0A0M3J1Y0 A0A2A6C9V2 A0A3P7S1D0 A0A0N4TEC4 A0A1I7ZNL3 A0A1I7ZPH5 A0A3P7E4K6 A0A368GXV9 A0A0K0FEA5 A0A0K0JE24 A0A0B2V3K6 A0A131YYX3 A0A2L2Y735 A0A182RDG1 B7P7B7 A0A147BD52 A0A0N4Y782 A0A087TK17 A0A2L2Y4K9
A0A0L7K498 A0A087UJV8 A0A0V1EPP9 A0A0V0Z7C1 A0A0V0S515 A0A2L2XYH3 A0A2L2XYN6 A0A182NIS5 A0A087TD78 B0XBB5 A0A1I7Z116 A0A2P6L3Q8 A0A1I7Z0X9 A0A0V0Z7M5 A0A1I7Z1E9 A0A0V1FGY8 A0A0V0Z791 A0A0V0S563 A0A0V0S5K9 T1L4R5 A0A2A4JLM8 A0A2P6KNP2 A0A0V1CXK4 A0A2Z6FL32 A0A194Q8M7 A0A194QS41 E5S2I0 A0A2P6L3P9 A0A0V0Z760 A0A087UDS0 A0A183J2C6 A0A0V0Z793 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1B9Y2 A0A0V1B9Z3 A0A0V1NT84 A0A0V0U5E1 A0A0V0WZ71 A0A0V1LSL0 A0A2H1WS62 A0A0V1H1U7 H9JHS7 A0A0V0U579 A0A0V0WZ93 A0A1I7ZRE6 A0A2P6L3P8 A0A0V1MZF6 A0A0V1EPP1 A0A0V1J3G8 A0A0V1J3E7 A0A0V1EPN2 E5T4Z6 A0A0N4X8U1 A0A1I8A6G1 A0A1Y3EJT5 A0A016VB53 A0A1I7YSE4 A0A0N5BI72 A0A2A2JIB4 A0A1I8A2H2 A0A0N5BYG0 A0A0M3J546 A0A0C2GUB0 A0A2P6LAR8 E7D188 A0A0D6LGI9 A0A016VAF8 A0A2P6L3T3 A0A2H1VNG5 A0A183UX28 D2A397 A0A2L2XZL2 A0A0L7LIA1 A0A0M3J1Y0 A0A2A6C9V2 A0A3P7S1D0 A0A0N4TEC4 A0A1I7ZNL3 A0A1I7ZPH5 A0A3P7E4K6 A0A368GXV9 A0A0K0FEA5 A0A0K0JE24 A0A0B2V3K6 A0A131YYX3 A0A2L2Y735 A0A182RDG1 B7P7B7 A0A147BD52 A0A0N4Y782 A0A087TK17 A0A2L2Y4K9
Ontologies
GO
PANTHER
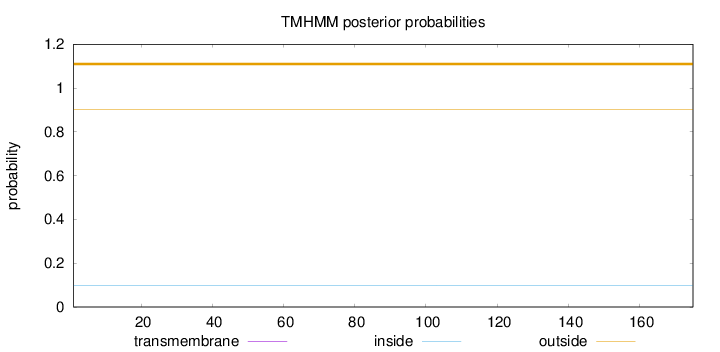
Topology
Length:
175
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09852
outside
1 - 175
Population Genetic Test Statistics
Pi
25.252441
Theta
211.174036
Tajima's D
1.780378
CLR
0.413939
CSRT
0.849407529623519
Interpretation
Uncertain
