Gene
KWMTBOMO01534
Pre Gene Modal
BGIBMGA009097
Annotation
PREDICTED:_N6-adenosine-methyltransferase_subunit_METTL14_[Amyelois_transitella]
Full name
N6-adenosine-methyltransferase non-catalytic subunit
Alternative Name
Karyogamy protein 4
Methyltransferase-like protein 14
Methyltransferase-like protein 14
Location in the cell
Extracellular Reliability : 3.891
Sequence
CDS
ATGAGTGAGAAATTAAGAGAACTACGCGAGAGATCTCAAAAACGAAAAAAGCTTCTAGCACAGACATTAGGTGTCTCGAGTGTGAGCGAATTAAGACATGCGCTGGGTTCAGCTCTAGACGTACCTGTTAAAAAACAAGCTGTGTCTGGAAGCGGAGGTGAAGGAGATAAACCCACCAGTAAGGACCAACCGGATGAACTTGTATACACCGACTCCTCTACATTTTTGAAGGGCACGCAGTCATCAAATCCTCACAATGACTATTGTCAGCACTTTGTTGACACAGGTCAGAGGCCACAAAACTATATCAGAGATGTTGGGCTCGCTGACAGGTTTGAGGAATATCCGAAATTACGTGAACTCATTAAACTGAAAGACGATTTAATTGCGGAAACTGCAACACCACCTATGTATTTGAAATGTGATCTCAAGACTTTTGATCTAAAAGACATGGCTAACAAATTCGATGTAATTCTCGTGGAACCTCCTCTGGGCACTGGATGGAGATGGGAAGATGTCTTAGCTCTTGAGCTCCAGCAGATAGCACAGCCAAGATCTTTTGTATTCCTATGGTGTGGAAGCTCTGAAGGTTTAGATATGGGCAGGGAATGCCTAAAGAAGTGGGGCTTTCGTCGTTGCGAAGACATCTGCTGGATCAAAACGAACATAAAGAATCCGGGGCATTCGAAAAACCTCGAGCACAACGCGGTCTTCCAGCGAACCAAGGAGCATTGTCTGATGGGCATCAAAGGCACTGTCCGGAGGTCGGTCGACGGAGACTTCATACACGCTAACGTGGATATCGACCTCATCATATCTGAAGACCCGGAGTTCGGCAGTACAGAGAAGCCTATTGAGATATTTCACATCATGGAACATTTCTGTTTGGGTCGAAGAAGGGTACACCTGTTCGGCAAGGACTCCACGATCCGTCCGGGCTGGGTGACGATCGGCCACGAGCTCACCAACTCCAACTTCAACGCGGAGCTGTACTCGTCGTACTTCGCCGAGGGCCGCGAGAGCACCGGCTGCACCGACAGGATCGAGGCCCTCAGGCCCAAGAGCCCCCCCAACACCAGCAAGGGCACCCGGGCCAGGGGCAGGGGCGGGTTTAGGGGCAGGGGGAGGGGCAGAGGGGCTATCTAA
Protein
MSEKLRELRERSQKRKKLLAQTLGVSSVSELRHALGSALDVPVKKQAVSGSGGEGDKPTSKDQPDELVYTDSSTFLKGTQSSNPHNDYCQHFVDTGQRPQNYIRDVGLADRFEEYPKLRELIKLKDDLIAETATPPMYLKCDLKTFDLKDMANKFDVILVEPPLGTGWRWEDVLALELQQIAQPRSFVFLWCGSSEGLDMGRECLKKWGFRRCEDICWIKTNIKNPGHSKNLEHNAVFQRTKEHCLMGIKGTVRRSVDGDFIHANVDIDLIISEDPEFGSTEKPIEIFHIMEHFCLGRRRVHLFGKDSTIRPGWVTIGHELTNSNFNAELYSSYFAEGRESTGCTDRIEALRPKSPPNTSKGTRARGRGGFRGRGRGRGAI
Summary
Description
Non-catalytic component of the WMM complex, a complex that mediates N6-methyladenosine (m6A) methylation of mRNAs, a modification that plays a role in the efficiency of mRNA splicing and is required for sex determination (PubMed:27919077, PubMed:28675155). In the heterodimer formed with Ime4/Mettl3, Mettl14 constitutes the RNA-binding scaffold that recognizes the substrate rather than the catalytic core (By similarity). Required for sex determination and dosage compensation via Sxl alternative splicing: m6A methylation acts as a key regulator of Sxl pre-mRNA and promotes female-specific alternative splicing of Sxl, which determines female physiognomy (PubMed:27919077, PubMed:28675155). M6A methylation is also required for neuronal functions (PubMed:27919077).
Subunit
Component of the WMM complex, a N6-methyltransferase complex composed of a catalytic subcomplex, named MAC, and of an associated subcomplex, named MACOM (PubMed:29535189, PubMed:28675155, PubMed:29555755). The MAC subcomplex is composed of Ime4/Mettl3 and Mettl14 (PubMed:29535189, PubMed:28675155, PubMed:29555755). The MACOM subcomplex is composed of fl(2)d, Flacc/Xio, Hakai, vir, and, in some cases of nito (PubMed:29535189, PubMed:29555755).
Similarity
Belongs to the MT-A70-like family.
Keywords
Complete proteome
Differentiation
mRNA processing
mRNA splicing
Nucleus
Reference proteome
RNA-binding
Sexual differentiation
Feature
chain N6-adenosine-methyltransferase non-catalytic subunit
Uniprot
H9JHU9
A0A2A4JE14
A0A2H1WD35
S4PPN1
A0A212EQ94
A0A194Q349
+ More
A0A1B6JJ16 A0A067R4L5 A0A1B6FC80 A0A2J7QGL2 A0A1B6MUY7 A0A0V0GAS6 T1IF26 A0A1W4WIV3 A0A069DT12 A0A1B6CCX0 A0A0T6BBE1 A0A1Y1KBE4 A0A0L7R541 A0A2A3EGX1 A0A088AW80 E0VZ87 A0A084WGV9 A0A182PMQ2 A0A1S3DF93 U5EWJ3 A0A023ERB4 F4WST8 A0A158NH17 A0A182VIQ9 A0A182TU14 A0A182WU37 A0A182L1H8 Q7Q3Z3 A0A026W1X5 A0A182HUP9 A0A182JK76 A0A182JQW9 A0A232FEV4 U4UDX9 A0A151J2F9 D6WV92 A0A195FN99 A0A151XDB5 A0A195B7B5 A0A310SBJ6 N6U3Q7 K7J3F6 A0A224XSA1 A0A1Q3F5M1 B0XBL4 A0A195D2S2 A0A182QI67 E9IMQ8 A0A0J7KXN8 A0A336MSY0 A0A336MRK6 A0A182NUC8 A0A182RZU5 A0A182M6Z5 A0A182W742 W8AGV8 W5J7V1 A0A2M4AM74 A0A182F4U6 A0A2M4BQL7 A0A182Y076 E2AJE3 T1PHX2 A0A0L0CDK7 A0A1I8Q120 A0A0A9XRN9 A0A1B0CZE0 A0A0K8UQI4 B4HYI2 A0A1W4UVI0 A0A1B0GCJ0 A0A0P4W8M7 Q9VLP7 D3TN32 A0A1B0BL96 A0A034WVG8 B4Q6Q0 A0A1B3PD81 A0A087UWN5 J9JVM4 B3N737 B4NWV8 A0A2H8TTH1 A0A2S2P3H8 A0A0N8AB70 Q29MN2 A0A3B0K1H3 A0A0P5MBK1 A0A2S2PYI1 A0A1L8DHF6 T1INW0 B4G902 A0A1A9Y3J6 B4MTJ3
A0A1B6JJ16 A0A067R4L5 A0A1B6FC80 A0A2J7QGL2 A0A1B6MUY7 A0A0V0GAS6 T1IF26 A0A1W4WIV3 A0A069DT12 A0A1B6CCX0 A0A0T6BBE1 A0A1Y1KBE4 A0A0L7R541 A0A2A3EGX1 A0A088AW80 E0VZ87 A0A084WGV9 A0A182PMQ2 A0A1S3DF93 U5EWJ3 A0A023ERB4 F4WST8 A0A158NH17 A0A182VIQ9 A0A182TU14 A0A182WU37 A0A182L1H8 Q7Q3Z3 A0A026W1X5 A0A182HUP9 A0A182JK76 A0A182JQW9 A0A232FEV4 U4UDX9 A0A151J2F9 D6WV92 A0A195FN99 A0A151XDB5 A0A195B7B5 A0A310SBJ6 N6U3Q7 K7J3F6 A0A224XSA1 A0A1Q3F5M1 B0XBL4 A0A195D2S2 A0A182QI67 E9IMQ8 A0A0J7KXN8 A0A336MSY0 A0A336MRK6 A0A182NUC8 A0A182RZU5 A0A182M6Z5 A0A182W742 W8AGV8 W5J7V1 A0A2M4AM74 A0A182F4U6 A0A2M4BQL7 A0A182Y076 E2AJE3 T1PHX2 A0A0L0CDK7 A0A1I8Q120 A0A0A9XRN9 A0A1B0CZE0 A0A0K8UQI4 B4HYI2 A0A1W4UVI0 A0A1B0GCJ0 A0A0P4W8M7 Q9VLP7 D3TN32 A0A1B0BL96 A0A034WVG8 B4Q6Q0 A0A1B3PD81 A0A087UWN5 J9JVM4 B3N737 B4NWV8 A0A2H8TTH1 A0A2S2P3H8 A0A0N8AB70 Q29MN2 A0A3B0K1H3 A0A0P5MBK1 A0A2S2PYI1 A0A1L8DHF6 T1INW0 B4G902 A0A1A9Y3J6 B4MTJ3
Pubmed
19121390
23622113
22118469
26354079
24845553
26334808
+ More
28004739 20566863 24438588 24945155 26483478 21719571 21347285 20966253 12364791 14747013 17210077 24508170 30249741 28648823 23537049 18362917 19820115 20075255 21282665 24495485 20920257 23761445 25244985 20798317 25315136 26108605 25401762 26823975 17994087 10731132 12537572 12537569 27919077 27919081 29535189 28675155 29555755 20353571 25348373 22936249 17550304 15632085
28004739 20566863 24438588 24945155 26483478 21719571 21347285 20966253 12364791 14747013 17210077 24508170 30249741 28648823 23537049 18362917 19820115 20075255 21282665 24495485 20920257 23761445 25244985 20798317 25315136 26108605 25401762 26823975 17994087 10731132 12537572 12537569 27919077 27919081 29535189 28675155 29555755 20353571 25348373 22936249 17550304 15632085
EMBL
BABH01029525
BABH01029526
BABH01029527
NWSH01001919
PCG69680.1
ODYU01007843
+ More
SOQ50973.1 GAIX01000147 JAA92413.1 AGBW02013278 OWR43662.1 KQ459562 KPI99768.1 GECU01008539 JAS99167.1 KK852699 KDR18181.1 GECZ01021988 JAS47781.1 NEVH01014361 PNF27717.1 GEBQ01000247 JAT39730.1 GECL01000940 JAP05184.1 ACPB03007378 GBGD01001799 JAC87090.1 GEDC01031023 GEDC01026047 JAS06275.1 JAS11251.1 LJIG01002312 KRT84628.1 GEZM01093889 JAV56087.1 KQ414654 KOC65944.1 KZ288256 PBC30532.1 DS235851 EEB18693.1 ATLV01023763 KE525346 KFB49453.1 GANO01001389 JAB58482.1 JXUM01071699 GAPW01001895 KQ562679 JAC11703.1 KXJ75339.1 GL888327 EGI62745.1 ADTU01015435 AAAB01008964 EAA12386.3 KK107487 QOIP01000006 EZA50070.1 RLU22167.1 APCN01002471 NNAY01000310 OXU29304.1 KB632099 ERL88796.1 KQ980447 KYN16013.1 KQ971363 EFA09090.2 KQ981387 KYN41968.1 KQ982294 KYQ58347.1 KQ976574 KYM80089.1 KQ761783 OAD57010.1 APGK01050753 KB741176 ENN73207.1 GFTR01005555 JAW10871.1 GFDL01012277 JAV22768.1 DS232641 EDS44326.1 KQ976973 KYN06664.1 AXCN02000765 GL764255 EFZ18091.1 LBMM01002075 KMQ95307.1 UFQS01002368 UFQT01002368 SSX13897.1 SSX33316.1 UFQS01001522 UFQT01001522 SSX11339.1 SSX30907.1 AXCM01002772 GAMC01021668 JAB84887.1 ADMH02002111 ETN58884.1 GGFK01008558 MBW41879.1 GGFJ01006236 MBW55377.1 GL439972 EFN66464.1 KA648412 AFP63041.1 JRES01000536 KNC30331.1 GBHO01020192 GBHO01020191 GBRD01011887 GDHC01017266 JAG23412.1 JAG23413.1 JAG53937.1 JAQ01363.1 AJVK01009544 GDHF01031244 GDHF01023566 JAI21070.1 JAI28748.1 CH480818 EDW52112.1 CCAG010021443 GDRN01081061 JAI62083.1 AE014134 AY069354 EZ422834 ADD19110.1 JXJN01016271 GAKP01000323 JAC58629.1 CM000361 CM002910 EDX04209.1 KMY89035.1 KU659872 AOG17671.1 KK122029 KFM81774.1 ABLF02012760 ABLF02012761 CH954177 EDV59333.1 CM000157 EDW88488.1 GFXV01005435 MBW17240.1 GGMR01011361 MBY23980.1 GDIP01164577 LRGB01000626 JAJ58825.1 KZS17486.1 CH379060 EAL33661.2 OUUW01000004 SPP79809.1 GDIQ01169672 JAK82053.1 GGMS01001374 MBY70577.1 GFDF01008310 JAV05774.1 JH431216 CH479180 EDW28832.1 CH963852 EDW75432.1
SOQ50973.1 GAIX01000147 JAA92413.1 AGBW02013278 OWR43662.1 KQ459562 KPI99768.1 GECU01008539 JAS99167.1 KK852699 KDR18181.1 GECZ01021988 JAS47781.1 NEVH01014361 PNF27717.1 GEBQ01000247 JAT39730.1 GECL01000940 JAP05184.1 ACPB03007378 GBGD01001799 JAC87090.1 GEDC01031023 GEDC01026047 JAS06275.1 JAS11251.1 LJIG01002312 KRT84628.1 GEZM01093889 JAV56087.1 KQ414654 KOC65944.1 KZ288256 PBC30532.1 DS235851 EEB18693.1 ATLV01023763 KE525346 KFB49453.1 GANO01001389 JAB58482.1 JXUM01071699 GAPW01001895 KQ562679 JAC11703.1 KXJ75339.1 GL888327 EGI62745.1 ADTU01015435 AAAB01008964 EAA12386.3 KK107487 QOIP01000006 EZA50070.1 RLU22167.1 APCN01002471 NNAY01000310 OXU29304.1 KB632099 ERL88796.1 KQ980447 KYN16013.1 KQ971363 EFA09090.2 KQ981387 KYN41968.1 KQ982294 KYQ58347.1 KQ976574 KYM80089.1 KQ761783 OAD57010.1 APGK01050753 KB741176 ENN73207.1 GFTR01005555 JAW10871.1 GFDL01012277 JAV22768.1 DS232641 EDS44326.1 KQ976973 KYN06664.1 AXCN02000765 GL764255 EFZ18091.1 LBMM01002075 KMQ95307.1 UFQS01002368 UFQT01002368 SSX13897.1 SSX33316.1 UFQS01001522 UFQT01001522 SSX11339.1 SSX30907.1 AXCM01002772 GAMC01021668 JAB84887.1 ADMH02002111 ETN58884.1 GGFK01008558 MBW41879.1 GGFJ01006236 MBW55377.1 GL439972 EFN66464.1 KA648412 AFP63041.1 JRES01000536 KNC30331.1 GBHO01020192 GBHO01020191 GBRD01011887 GDHC01017266 JAG23412.1 JAG23413.1 JAG53937.1 JAQ01363.1 AJVK01009544 GDHF01031244 GDHF01023566 JAI21070.1 JAI28748.1 CH480818 EDW52112.1 CCAG010021443 GDRN01081061 JAI62083.1 AE014134 AY069354 EZ422834 ADD19110.1 JXJN01016271 GAKP01000323 JAC58629.1 CM000361 CM002910 EDX04209.1 KMY89035.1 KU659872 AOG17671.1 KK122029 KFM81774.1 ABLF02012760 ABLF02012761 CH954177 EDV59333.1 CM000157 EDW88488.1 GFXV01005435 MBW17240.1 GGMR01011361 MBY23980.1 GDIP01164577 LRGB01000626 JAJ58825.1 KZS17486.1 CH379060 EAL33661.2 OUUW01000004 SPP79809.1 GDIQ01169672 JAK82053.1 GGMS01001374 MBY70577.1 GFDF01008310 JAV05774.1 JH431216 CH479180 EDW28832.1 CH963852 EDW75432.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000027135
UP000235965
+ More
UP000015103 UP000192223 UP000053825 UP000242457 UP000005203 UP000009046 UP000030765 UP000075885 UP000079169 UP000069940 UP000249989 UP000007755 UP000005205 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000053097 UP000279307 UP000075840 UP000075880 UP000075881 UP000215335 UP000030742 UP000078492 UP000007266 UP000078541 UP000075809 UP000078540 UP000019118 UP000002358 UP000002320 UP000078542 UP000075886 UP000036403 UP000075884 UP000075900 UP000075883 UP000075920 UP000000673 UP000069272 UP000076408 UP000000311 UP000095301 UP000037069 UP000095300 UP000092462 UP000001292 UP000192221 UP000092444 UP000000803 UP000092460 UP000000304 UP000054359 UP000007819 UP000008711 UP000002282 UP000076858 UP000001819 UP000268350 UP000008744 UP000092443 UP000007798
UP000015103 UP000192223 UP000053825 UP000242457 UP000005203 UP000009046 UP000030765 UP000075885 UP000079169 UP000069940 UP000249989 UP000007755 UP000005205 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000053097 UP000279307 UP000075840 UP000075880 UP000075881 UP000215335 UP000030742 UP000078492 UP000007266 UP000078541 UP000075809 UP000078540 UP000019118 UP000002358 UP000002320 UP000078542 UP000075886 UP000036403 UP000075884 UP000075900 UP000075883 UP000075920 UP000000673 UP000069272 UP000076408 UP000000311 UP000095301 UP000037069 UP000095300 UP000092462 UP000001292 UP000192221 UP000092444 UP000000803 UP000092460 UP000000304 UP000054359 UP000007819 UP000008711 UP000002282 UP000076858 UP000001819 UP000268350 UP000008744 UP000092443 UP000007798
Pfam
PF05063 MT-A70
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JHU9
A0A2A4JE14
A0A2H1WD35
S4PPN1
A0A212EQ94
A0A194Q349
+ More
A0A1B6JJ16 A0A067R4L5 A0A1B6FC80 A0A2J7QGL2 A0A1B6MUY7 A0A0V0GAS6 T1IF26 A0A1W4WIV3 A0A069DT12 A0A1B6CCX0 A0A0T6BBE1 A0A1Y1KBE4 A0A0L7R541 A0A2A3EGX1 A0A088AW80 E0VZ87 A0A084WGV9 A0A182PMQ2 A0A1S3DF93 U5EWJ3 A0A023ERB4 F4WST8 A0A158NH17 A0A182VIQ9 A0A182TU14 A0A182WU37 A0A182L1H8 Q7Q3Z3 A0A026W1X5 A0A182HUP9 A0A182JK76 A0A182JQW9 A0A232FEV4 U4UDX9 A0A151J2F9 D6WV92 A0A195FN99 A0A151XDB5 A0A195B7B5 A0A310SBJ6 N6U3Q7 K7J3F6 A0A224XSA1 A0A1Q3F5M1 B0XBL4 A0A195D2S2 A0A182QI67 E9IMQ8 A0A0J7KXN8 A0A336MSY0 A0A336MRK6 A0A182NUC8 A0A182RZU5 A0A182M6Z5 A0A182W742 W8AGV8 W5J7V1 A0A2M4AM74 A0A182F4U6 A0A2M4BQL7 A0A182Y076 E2AJE3 T1PHX2 A0A0L0CDK7 A0A1I8Q120 A0A0A9XRN9 A0A1B0CZE0 A0A0K8UQI4 B4HYI2 A0A1W4UVI0 A0A1B0GCJ0 A0A0P4W8M7 Q9VLP7 D3TN32 A0A1B0BL96 A0A034WVG8 B4Q6Q0 A0A1B3PD81 A0A087UWN5 J9JVM4 B3N737 B4NWV8 A0A2H8TTH1 A0A2S2P3H8 A0A0N8AB70 Q29MN2 A0A3B0K1H3 A0A0P5MBK1 A0A2S2PYI1 A0A1L8DHF6 T1INW0 B4G902 A0A1A9Y3J6 B4MTJ3
A0A1B6JJ16 A0A067R4L5 A0A1B6FC80 A0A2J7QGL2 A0A1B6MUY7 A0A0V0GAS6 T1IF26 A0A1W4WIV3 A0A069DT12 A0A1B6CCX0 A0A0T6BBE1 A0A1Y1KBE4 A0A0L7R541 A0A2A3EGX1 A0A088AW80 E0VZ87 A0A084WGV9 A0A182PMQ2 A0A1S3DF93 U5EWJ3 A0A023ERB4 F4WST8 A0A158NH17 A0A182VIQ9 A0A182TU14 A0A182WU37 A0A182L1H8 Q7Q3Z3 A0A026W1X5 A0A182HUP9 A0A182JK76 A0A182JQW9 A0A232FEV4 U4UDX9 A0A151J2F9 D6WV92 A0A195FN99 A0A151XDB5 A0A195B7B5 A0A310SBJ6 N6U3Q7 K7J3F6 A0A224XSA1 A0A1Q3F5M1 B0XBL4 A0A195D2S2 A0A182QI67 E9IMQ8 A0A0J7KXN8 A0A336MSY0 A0A336MRK6 A0A182NUC8 A0A182RZU5 A0A182M6Z5 A0A182W742 W8AGV8 W5J7V1 A0A2M4AM74 A0A182F4U6 A0A2M4BQL7 A0A182Y076 E2AJE3 T1PHX2 A0A0L0CDK7 A0A1I8Q120 A0A0A9XRN9 A0A1B0CZE0 A0A0K8UQI4 B4HYI2 A0A1W4UVI0 A0A1B0GCJ0 A0A0P4W8M7 Q9VLP7 D3TN32 A0A1B0BL96 A0A034WVG8 B4Q6Q0 A0A1B3PD81 A0A087UWN5 J9JVM4 B3N737 B4NWV8 A0A2H8TTH1 A0A2S2P3H8 A0A0N8AB70 Q29MN2 A0A3B0K1H3 A0A0P5MBK1 A0A2S2PYI1 A0A1L8DHF6 T1INW0 B4G902 A0A1A9Y3J6 B4MTJ3
PDB
5TEY
E-value=1.33303e-136,
Score=1246
Ontologies
GO
Topology
Subcellular location
Nucleus
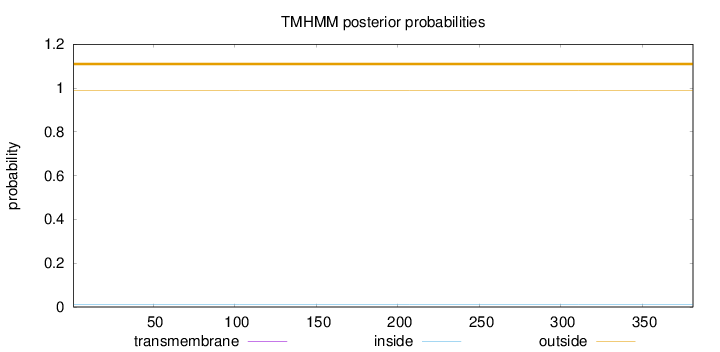
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00117
Exp number, first 60 AAs:
0.00117
Total prob of N-in:
0.01248
outside
1 - 381
Population Genetic Test Statistics
Pi
368.548902
Theta
176.825143
Tajima's D
3.417066
CLR
0.369437
CSRT
0.992700364981751
Interpretation
Uncertain
