Gene
KWMTBOMO01533
Pre Gene Modal
BGIBMGA009071
Annotation
PREDICTED:_phosphatidylinositol_3?4?5-trisphosphate_3-phosphatase_and_dual-specificity_protein_phosphatase_PTEN_isoform_X1_[Bombyx_mori]
Full name
Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN
Alternative Name
Phosphatase and tensin homolog
Location in the cell
Nuclear Reliability : 2.484
Sequence
CDS
ATGGGTGTTTGCGTGAGCTGCCGGCGCCGAGAGCGGAAGATCCCAGAAAAGCGGTCGGTGCCGGGAGTGGTTGGGCCCGCTAAGGGCGCGGCGGCGGAGCTGCTGCCCAGGCCAGTGAATCGCTTTACAGCCATGGCAAACTCCATGTCAAATATCAAGATGACCAACCCGATAAAGAATATGGTCAGCAAGCGCAGGATACGCTACACCAAGCACGGATTCAACCTGGACTTAGCCTATATCACAGACAGGCTGATAGCGATGGGTTTCCCGGCCGAGAAATTGGAAGGAGTATACAGGAATCACATAGACGAAGTGTATAGGTTTTTAGAGACAATGCACAAGGATCACTACAAGATATACAACTTGTGCTCCGAGAGGACATACGACCCGAGCAAGTTTCATAAGAGGGTGGAGAGATATGCGTTCGAAGACCACACCCCGCCCCACATGGAGCTGATACAGCCATTCTGCGAGAACGTGCACAGTTGGCTCAGTGCGGATACAAGGAACGTTGCCGTCGTACATTGTAAAGCGGGGAAAGGTCGGACCGGTACAATGGTCTGTTGTTATTTGTTATACAGCGGGGACCAGCCTAATGCAGACGAAGCGCTCAAGTTCTACGGCAGGAAGAGAACTCTAGATGAAAAGGGCGTGACGATACCGTCGCAGCGGCGCTACGTGGAGTACTACGGGGCGCTGCTGCGGTCGGGCGTGGCGTACCGGGCGGGCGCGGCGCGCGTGCAGGTCCGCGAGCTGCGCATGTCGCCGCCGCCCGCCGCCCACGTGCCCGCCGCCGCCGCGCCGCCCGACCTCGTGCTGTGCCAGGCCGACCCGCACGCCGACCCGCACAGCGAACACAACGTGCTCGTGCTCAAGACGCCGCCCGGATGCGTGGATGTGCAGCGAGACGAGAACAGCATCCGCATCTGCCTGACGCAGTGCACGCCGCTGTTCGGCGACATCAGGGTGGACGTCTACAACAAACCGAAGATGAAGATGCGCAAAGAGAAGCTGTTTCATTTCTGGTTCAACACGTTCTTCGTTACGGCCGAAGTCGGAGCCCAAAGAATACCGCCATTACCTGACAGTCCGGACCTGGAGACGTACAAGCTGACCCTGAACAAGTGGCAGCTGGACGACGCCCACAAGGACAAGCAACACAAACATTACAGCCCCGACTTCAAGGTTGAGCTGATAGTGGAGAAGGTGCCGGATTCGTCGACGTACAGACCGGCGTGCGACCGCACCAGGTCACCGACCTCCTCGTCCTCCTCCTCCGGCTCCGTCGGCTCCGACCCCGAGACGGACGGGAACTGGGACTCTGGGACGGTGTGCGAGCGCGTCGGCCATTACCGGCAGCTGTCCCCCGACCACACGTGA
Protein
MGVCVSCRRRERKIPEKRSVPGVVGPAKGAAAELLPRPVNRFTAMANSMSNIKMTNPIKNMVSKRRIRYTKHGFNLDLAYITDRLIAMGFPAEKLEGVYRNHIDEVYRFLETMHKDHYKIYNLCSERTYDPSKFHKRVERYAFEDHTPPHMELIQPFCENVHSWLSADTRNVAVVHCKAGKGRTGTMVCCYLLYSGDQPNADEALKFYGRKRTLDEKGVTIPSQRRYVEYYGALLRSGVAYRAGAARVQVRELRMSPPPAAHVPAAAAPPDLVLCQADPHADPHSEHNVLVLKTPPGCVDVQRDENSIRICLTQCTPLFGDIRVDVYNKPKMKMRKEKLFHFWFNTFFVTAEVGAQRIPPLPDSPDLETYKLTLNKWQLDDAHKDKQHKHYSPDFKVELIVEKVPDSSTYRPACDRTRSPTSSSSSSGSVGSDPETDGNWDSGTVCERVGHYRQLSPDHT
Summary
Description
Tumor suppressor. Acts as a dual-specificity protein phosphatase, dephosphorylating tyrosine-, serine- and threonine-phosphorylated proteins. Also acts as a lipid phosphatase.
Catalytic Activity
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4,5-trisphosphate) + H2O = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4,5-trisphosphate) + H2O = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + phosphate
Similarity
Belongs to the PTEN phosphatase protein family.
Uniprot
A0A1U9VYP0
A0A385KP72
A0A194Q425
A0A212EQA5
A0A2A4JC56
A0A2J7PXJ8
+ More
A0A2J7PXI9 A0A2J7PXJ5 A0A1B6IPR6 A0A1B6I2P7 A0A1B6E296 A0A1B6HBA5 A0A1B6FF62 A0A1W4W8J9 A0A1W4WJU2 A0A026W202 T1IEZ7 A0A3L8DNZ8 A0A1W4WK32 E2B2C4 K7J3F7 C5IX04 C5IX06 A0A2A3EHP8 A0A088AW82 A0A310SNU5 A0A195B6F6 A0A158NH16 A0A151J1Z4 C5IX05 A0A195D1D6 A0A1Y1NNP8 A0A1Y1NIW4 A0A1Y1NMX4 A0A0C9QNZ4 A0A232FEW7 A0A195FPW0 A0A0C9REB5 A0A224XCD3 A0A1L8DKY8 A0A0C9RAI0 A0A023F2V6 A0A067R4W3 A0A0V0GEQ0 V5I9N9 V5G6Z5 A0A139WCR0 U5EGU8 D6WV93 E9IMQ7 V5GVX9 A0A0A9Z3S5 A0A0A9ZDM8 A0A146KZH4 A0A0A9WR37 A0A0A9ZDN1 E2AJE6 A0A0J7KUV5 A0A336LPS5 R7UIY8 A0A336LL10 A0A3Q0IHJ6 C3Y027 A0A0M5L082 A0A1B0GFE4 A0A2S2PGK5 J9JVA7 A0A0T6B671 A0A182YYW0 A0A2H8TZB0 A0A1A9WHF5 A0A2S2QLC3 A0A1I8MDR5 W8BRK9 A0A1B0AH81 A0A1B0C653 A0A1A9Y710 A0A1A9UD18 A0A1S3IBE2 A0A1I8NP83 A0A1W4USY4 A0A147AZ98 F7EQJ0 A0A1W4W8K1 A0A0A1X1W9 I0BWL0 L8HXB3 W5PXK2 A0A0K8VMJ7 A1Z072 A1Z068 A0A1S2ZKK5 I3MU41 A0A2K6EIG7
A0A2J7PXI9 A0A2J7PXJ5 A0A1B6IPR6 A0A1B6I2P7 A0A1B6E296 A0A1B6HBA5 A0A1B6FF62 A0A1W4W8J9 A0A1W4WJU2 A0A026W202 T1IEZ7 A0A3L8DNZ8 A0A1W4WK32 E2B2C4 K7J3F7 C5IX04 C5IX06 A0A2A3EHP8 A0A088AW82 A0A310SNU5 A0A195B6F6 A0A158NH16 A0A151J1Z4 C5IX05 A0A195D1D6 A0A1Y1NNP8 A0A1Y1NIW4 A0A1Y1NMX4 A0A0C9QNZ4 A0A232FEW7 A0A195FPW0 A0A0C9REB5 A0A224XCD3 A0A1L8DKY8 A0A0C9RAI0 A0A023F2V6 A0A067R4W3 A0A0V0GEQ0 V5I9N9 V5G6Z5 A0A139WCR0 U5EGU8 D6WV93 E9IMQ7 V5GVX9 A0A0A9Z3S5 A0A0A9ZDM8 A0A146KZH4 A0A0A9WR37 A0A0A9ZDN1 E2AJE6 A0A0J7KUV5 A0A336LPS5 R7UIY8 A0A336LL10 A0A3Q0IHJ6 C3Y027 A0A0M5L082 A0A1B0GFE4 A0A2S2PGK5 J9JVA7 A0A0T6B671 A0A182YYW0 A0A2H8TZB0 A0A1A9WHF5 A0A2S2QLC3 A0A1I8MDR5 W8BRK9 A0A1B0AH81 A0A1B0C653 A0A1A9Y710 A0A1A9UD18 A0A1S3IBE2 A0A1I8NP83 A0A1W4USY4 A0A147AZ98 F7EQJ0 A0A1W4W8K1 A0A0A1X1W9 I0BWL0 L8HXB3 W5PXK2 A0A0K8VMJ7 A1Z072 A1Z068 A0A1S2ZKK5 I3MU41 A0A2K6EIG7
EC Number
3.1.3.16
3.1.3.48
3.1.3.67
3.1.3.48
3.1.3.67
Pubmed
EMBL
KX792108
AQY15761.1
MG657022
AXZ96482.1
KQ459562
KPI99764.1
+ More
AGBW02013278 OWR43665.1 NWSH01001919 PCG69677.1 NEVH01020856 PNF21052.1 PNF21051.1 PNF21053.1 GECU01018813 JAS88893.1 GECU01026571 JAS81135.1 GEDC01031216 GEDC01025579 GEDC01021541 GEDC01008224 GEDC01005281 JAS06082.1 JAS11719.1 JAS15757.1 JAS29074.1 JAS32017.1 GECU01035776 GECU01003138 JAS71930.1 JAT04569.1 GECZ01020942 JAS48827.1 KK107487 EZA50068.1 ACPB03007378 QOIP01000006 RLU22170.1 GL445130 EFN90161.1 FJ969918 ACR55735.1 FJ969920 ACR55737.1 KZ288256 PBC30531.1 KQ761783 OAD57012.1 KQ976574 KYM80091.1 ADTU01015432 KQ980447 KYN16012.1 FJ969919 ACR55736.1 KQ976973 KYN06661.1 GEZM01001533 JAV97697.1 GEZM01001531 JAV97699.1 GEZM01001534 JAV97696.1 GBYB01005269 JAG75036.1 NNAY01000310 OXU29306.1 KQ981387 KYN41969.1 GBYB01005268 GBYB01005271 JAG75035.1 JAG75038.1 GFTR01006379 JAW10047.1 GFDF01006941 JAV07143.1 GBYB01005270 JAG75037.1 GBBI01003174 JAC15538.1 KK852699 KDR18177.1 GECL01000386 JAP05738.1 GALX01002605 JAB65861.1 GALX01002606 JAB65860.1 KQ971363 KYB25713.1 GANO01003297 JAB56574.1 EFA07760.2 GL764255 EFZ18116.1 GALX01002604 JAB65862.1 GBHO01003683 GBHO01003682 GBRD01017941 GBRD01017940 GDHC01000972 JAG39921.1 JAG39922.1 JAG47886.1 JAQ17657.1 GBHO01003684 GBHO01003681 JAG39920.1 JAG39923.1 GDHC01017440 JAQ01189.1 GBHO01033718 GBHO01033717 GBHO01021857 GBHO01003686 JAG09886.1 JAG09887.1 JAG21747.1 JAG39918.1 GBHO01003685 GBHO01003679 GBRD01017939 GBRD01017938 GDHC01010648 GDHC01004693 JAG39919.1 JAG39925.1 JAG47888.1 JAQ07981.1 JAQ13936.1 GL439972 EFN66467.1 LBMM01003020 KMQ94063.1 UFQT01000037 SSX18397.1 AMQN01007375 KB300556 ELU06519.1 SSX18395.1 GG666477 EEN66393.1 KR075846 ALE20565.1 CCAG010014184 GGMR01015950 MBY28569.1 ABLF02006459 LJIG01009534 KRT82888.1 GFXV01007772 MBW19577.1 GGMS01009344 MBY78547.1 GAMC01004773 JAC01783.1 JXJN01026447 GCES01002440 JAR83883.1 GBXI01008993 JAD05299.1 JQ326290 AFH66808.1 JH882713 ELR48481.1 AMGL01063172 AMGL01063173 AMGL01063174 GDHF01012210 JAI40104.1 EF153770 ABM21568.1 EF153766 ABM21564.1 AGTP01101032 AGTP01101033
AGBW02013278 OWR43665.1 NWSH01001919 PCG69677.1 NEVH01020856 PNF21052.1 PNF21051.1 PNF21053.1 GECU01018813 JAS88893.1 GECU01026571 JAS81135.1 GEDC01031216 GEDC01025579 GEDC01021541 GEDC01008224 GEDC01005281 JAS06082.1 JAS11719.1 JAS15757.1 JAS29074.1 JAS32017.1 GECU01035776 GECU01003138 JAS71930.1 JAT04569.1 GECZ01020942 JAS48827.1 KK107487 EZA50068.1 ACPB03007378 QOIP01000006 RLU22170.1 GL445130 EFN90161.1 FJ969918 ACR55735.1 FJ969920 ACR55737.1 KZ288256 PBC30531.1 KQ761783 OAD57012.1 KQ976574 KYM80091.1 ADTU01015432 KQ980447 KYN16012.1 FJ969919 ACR55736.1 KQ976973 KYN06661.1 GEZM01001533 JAV97697.1 GEZM01001531 JAV97699.1 GEZM01001534 JAV97696.1 GBYB01005269 JAG75036.1 NNAY01000310 OXU29306.1 KQ981387 KYN41969.1 GBYB01005268 GBYB01005271 JAG75035.1 JAG75038.1 GFTR01006379 JAW10047.1 GFDF01006941 JAV07143.1 GBYB01005270 JAG75037.1 GBBI01003174 JAC15538.1 KK852699 KDR18177.1 GECL01000386 JAP05738.1 GALX01002605 JAB65861.1 GALX01002606 JAB65860.1 KQ971363 KYB25713.1 GANO01003297 JAB56574.1 EFA07760.2 GL764255 EFZ18116.1 GALX01002604 JAB65862.1 GBHO01003683 GBHO01003682 GBRD01017941 GBRD01017940 GDHC01000972 JAG39921.1 JAG39922.1 JAG47886.1 JAQ17657.1 GBHO01003684 GBHO01003681 JAG39920.1 JAG39923.1 GDHC01017440 JAQ01189.1 GBHO01033718 GBHO01033717 GBHO01021857 GBHO01003686 JAG09886.1 JAG09887.1 JAG21747.1 JAG39918.1 GBHO01003685 GBHO01003679 GBRD01017939 GBRD01017938 GDHC01010648 GDHC01004693 JAG39919.1 JAG39925.1 JAG47888.1 JAQ07981.1 JAQ13936.1 GL439972 EFN66467.1 LBMM01003020 KMQ94063.1 UFQT01000037 SSX18397.1 AMQN01007375 KB300556 ELU06519.1 SSX18395.1 GG666477 EEN66393.1 KR075846 ALE20565.1 CCAG010014184 GGMR01015950 MBY28569.1 ABLF02006459 LJIG01009534 KRT82888.1 GFXV01007772 MBW19577.1 GGMS01009344 MBY78547.1 GAMC01004773 JAC01783.1 JXJN01026447 GCES01002440 JAR83883.1 GBXI01008993 JAD05299.1 JQ326290 AFH66808.1 JH882713 ELR48481.1 AMGL01063172 AMGL01063173 AMGL01063174 GDHF01012210 JAI40104.1 EF153770 ABM21568.1 EF153766 ABM21564.1 AGTP01101032 AGTP01101033
Proteomes
UP000053268
UP000007151
UP000218220
UP000235965
UP000192223
UP000053097
+ More
UP000015103 UP000279307 UP000008237 UP000002358 UP000242457 UP000005203 UP000078540 UP000005205 UP000078492 UP000078542 UP000215335 UP000078541 UP000027135 UP000007266 UP000000311 UP000036403 UP000014760 UP000079169 UP000001554 UP000092444 UP000007819 UP000076420 UP000091820 UP000095301 UP000092445 UP000092460 UP000092443 UP000078200 UP000085678 UP000095300 UP000192221 UP000265000 UP000002280 UP000002356 UP000079721 UP000005215 UP000233160
UP000015103 UP000279307 UP000008237 UP000002358 UP000242457 UP000005203 UP000078540 UP000005205 UP000078492 UP000078542 UP000215335 UP000078541 UP000027135 UP000007266 UP000000311 UP000036403 UP000014760 UP000079169 UP000001554 UP000092444 UP000007819 UP000076420 UP000091820 UP000095301 UP000092445 UP000092460 UP000092443 UP000078200 UP000085678 UP000095300 UP000192221 UP000265000 UP000002280 UP000002356 UP000079721 UP000005215 UP000233160
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
A0A1U9VYP0
A0A385KP72
A0A194Q425
A0A212EQA5
A0A2A4JC56
A0A2J7PXJ8
+ More
A0A2J7PXI9 A0A2J7PXJ5 A0A1B6IPR6 A0A1B6I2P7 A0A1B6E296 A0A1B6HBA5 A0A1B6FF62 A0A1W4W8J9 A0A1W4WJU2 A0A026W202 T1IEZ7 A0A3L8DNZ8 A0A1W4WK32 E2B2C4 K7J3F7 C5IX04 C5IX06 A0A2A3EHP8 A0A088AW82 A0A310SNU5 A0A195B6F6 A0A158NH16 A0A151J1Z4 C5IX05 A0A195D1D6 A0A1Y1NNP8 A0A1Y1NIW4 A0A1Y1NMX4 A0A0C9QNZ4 A0A232FEW7 A0A195FPW0 A0A0C9REB5 A0A224XCD3 A0A1L8DKY8 A0A0C9RAI0 A0A023F2V6 A0A067R4W3 A0A0V0GEQ0 V5I9N9 V5G6Z5 A0A139WCR0 U5EGU8 D6WV93 E9IMQ7 V5GVX9 A0A0A9Z3S5 A0A0A9ZDM8 A0A146KZH4 A0A0A9WR37 A0A0A9ZDN1 E2AJE6 A0A0J7KUV5 A0A336LPS5 R7UIY8 A0A336LL10 A0A3Q0IHJ6 C3Y027 A0A0M5L082 A0A1B0GFE4 A0A2S2PGK5 J9JVA7 A0A0T6B671 A0A182YYW0 A0A2H8TZB0 A0A1A9WHF5 A0A2S2QLC3 A0A1I8MDR5 W8BRK9 A0A1B0AH81 A0A1B0C653 A0A1A9Y710 A0A1A9UD18 A0A1S3IBE2 A0A1I8NP83 A0A1W4USY4 A0A147AZ98 F7EQJ0 A0A1W4W8K1 A0A0A1X1W9 I0BWL0 L8HXB3 W5PXK2 A0A0K8VMJ7 A1Z072 A1Z068 A0A1S2ZKK5 I3MU41 A0A2K6EIG7
A0A2J7PXI9 A0A2J7PXJ5 A0A1B6IPR6 A0A1B6I2P7 A0A1B6E296 A0A1B6HBA5 A0A1B6FF62 A0A1W4W8J9 A0A1W4WJU2 A0A026W202 T1IEZ7 A0A3L8DNZ8 A0A1W4WK32 E2B2C4 K7J3F7 C5IX04 C5IX06 A0A2A3EHP8 A0A088AW82 A0A310SNU5 A0A195B6F6 A0A158NH16 A0A151J1Z4 C5IX05 A0A195D1D6 A0A1Y1NNP8 A0A1Y1NIW4 A0A1Y1NMX4 A0A0C9QNZ4 A0A232FEW7 A0A195FPW0 A0A0C9REB5 A0A224XCD3 A0A1L8DKY8 A0A0C9RAI0 A0A023F2V6 A0A067R4W3 A0A0V0GEQ0 V5I9N9 V5G6Z5 A0A139WCR0 U5EGU8 D6WV93 E9IMQ7 V5GVX9 A0A0A9Z3S5 A0A0A9ZDM8 A0A146KZH4 A0A0A9WR37 A0A0A9ZDN1 E2AJE6 A0A0J7KUV5 A0A336LPS5 R7UIY8 A0A336LL10 A0A3Q0IHJ6 C3Y027 A0A0M5L082 A0A1B0GFE4 A0A2S2PGK5 J9JVA7 A0A0T6B671 A0A182YYW0 A0A2H8TZB0 A0A1A9WHF5 A0A2S2QLC3 A0A1I8MDR5 W8BRK9 A0A1B0AH81 A0A1B0C653 A0A1A9Y710 A0A1A9UD18 A0A1S3IBE2 A0A1I8NP83 A0A1W4USY4 A0A147AZ98 F7EQJ0 A0A1W4W8K1 A0A0A1X1W9 I0BWL0 L8HXB3 W5PXK2 A0A0K8VMJ7 A1Z072 A1Z068 A0A1S2ZKK5 I3MU41 A0A2K6EIG7
PDB
1D5R
E-value=8.65549e-80,
Score=757
Ontologies
PATHWAY
00562
Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04070 Phosphatidylinositol signaling system - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04070 Phosphatidylinositol signaling system - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
GO
GO:0008138
GO:0004725
GO:0016301
GO:0016314
GO:0051800
GO:0046855
GO:0051717
GO:0046856
GO:0008285
GO:0007399
GO:0016605
GO:0005737
GO:0004438
GO:0019901
GO:0042711
GO:0060736
GO:0008284
GO:0002902
GO:0030165
GO:0005829
GO:0032228
GO:0060997
GO:0033555
GO:0070373
GO:0043066
GO:0004722
GO:0071257
GO:0045736
GO:0060044
GO:0060291
GO:1903690
GO:0021955
GO:0090344
GO:0032286
GO:0060070
GO:0070374
GO:0097107
GO:0051898
GO:0001525
GO:0010719
GO:0051895
GO:0030534
GO:0043491
GO:0060179
GO:0016324
GO:0051091
GO:0050680
GO:2000463
GO:0009898
GO:0090071
GO:0060024
GO:0048738
GO:0043542
GO:0050821
GO:0045475
GO:0007270
GO:0051548
GO:0033032
GO:0035749
GO:0090394
GO:2000808
GO:0007611
GO:0043220
GO:1903984
GO:0060134
GO:0031642
GO:0071456
GO:0043005
GO:0097105
GO:0021542
GO:1990381
GO:0048853
GO:0042802
GO:0035176
GO:0060074
GO:0045792
GO:2000060
GO:0050771
GO:0061002
GO:1904668
GO:0010997
GO:0048681
GO:2000134
GO:1904706
GO:0006470
GO:0005515
GO:0016311
GO:0016791
GO:0006418
GO:0006334
GO:0003779
GO:0016021
Topology
Subcellular location
Cytoplasm
Nucleus
PML body
Nucleus
PML body
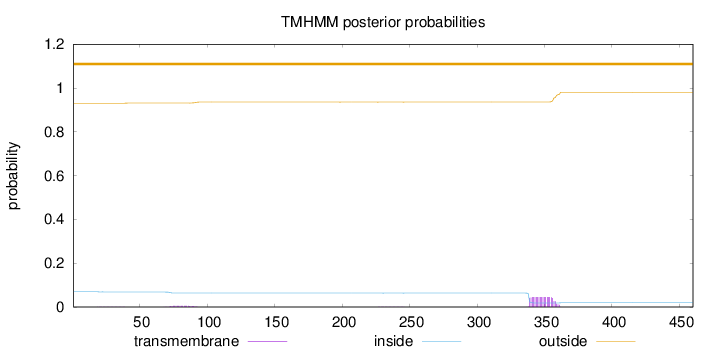
Length:
460
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.01085
Exp number, first 60 AAs:
0.01366
Total prob of N-in:
0.06955
outside
1 - 460
Population Genetic Test Statistics
Pi
234.838654
Theta
164.798082
Tajima's D
1.294244
CLR
0.339883
CSRT
0.733513324333783
Interpretation
Uncertain
