Pre Gene Modal
BGIBMGA009099
Annotation
PREDICTED:_ATPase_family_AAA_domain-containing_protein_1-like_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.822 Mitochondrial Reliability : 1.308
Sequence
CDS
ATGCCCGCCACCTTCCACGTGCCCATGCCCAGCGAGATGCAACGGGAACGGATACTCAAACTCATCCTGCAATCCGAGCCGGTTTCTGATGATATCGAGTACCGGCGGCTGGCGGCGGCGACCGACGGCTTCTCGGGCTCCGACCTGCACGAGCTGTGCCGCCACGCCGCCGTGTACCGCGTGCGGGACCTCGCAAGGGACGAGGTGGCCAAGGAGATGAACCAGAAGGAGAACCGGTCGAGTACGGAGTCGGACGACGAGTACCAGGACGCGGTCCGGCCCATCACGATGGAGGACTTGCGTTTGTCGCTCAACAAACTGAAGGAGTCCAAGATACAGTGCGGCGCCCTGCCCGGACACGTGAGGATAGAGCTCGACTAG
Protein
MPATFHVPMPSEMQRERILKLILQSEPVSDDIEYRRLAAATDGFSGSDLHELCRHAAVYRVRDLARDEVAKEMNQKENRSSTESDDEYQDAVRPITMEDLRLSLNKLKESKIQCGALPGHVRIELD
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9JHV1
A0A2A4J8M5
A0A2A4J7Q1
A0A194Q2H7
A0A194QR36
A0A212EQA7
+ More
S4PN20 A0A1Y1MSL6 D6WV96 N6TVA1 A0A1W4WIT2 V9IIG1 A0A2A3EFN4 A0A088AW81 A0A232FG58 A0A067R6Z3 A0A026W1X4 A0A310SPY8 A0A0L7R5A7 E9IMQ6 A0A151XD95 A0A2J7PXJ2 A0A1B6JS33 A0A195D173 A0A0V0G4P5 A0A2P8YIF9 A0A069DTJ3 A0A1Z5L2Y9 A0A195FMY5 A0A158NH14 F4WST6 A0A2R5LGM7 A0A151J1Z5 A0A1B6GM89 A0A1B6KGF7 A0A131Y1S6 A0A1B6HSL5 A0A293MZW9 R4WD53 A0A0P4VS30 R4FLB3 E2AJE7 A0A0J7KUG9 W8CAD1 A0A1B6DC69 A0A0T6BEB5 A0A293LJM6 A0A0K8R5Z9 E2B2C5 A0A195B759 A0A1B0BLN9 A0A1A9XJC8 A0A0A1WL38 A0A3P9Q4X7 A0A3B3VP53 A0A2L2Y073 A0A0K8WJD2 A0A034WAT0 A0A0M3QTM9 A0A131XDK6 A0A3B3YZR5 A0A1E1XCS4 A0A087Y215 A0A2R7WWI9 A0A131YIZ0 A0A224YRS1 A0A0A9YZ97 A0A1A9URM4 A0A146M1E4 A0A1A9ZVR1 B4JP30 L7M000 B4Q8T1 A0A023GP71 A0A0J9R0V6 B4KJG7 A0A023FLI3 A0A0C9SE65 D3TNG6 A0A1A9WWC6 A0A3B3QUA3 X2J7P4 M4AU55 Q9VL02 A0A0K8TS53 A0A1L8E0D6 A0A1L8E056 B4HWK0 B4MUD1 B3MPQ5 B3N965 A0A1B0D1U9 B4G777 B4M8G5 B4NZ89 U5EKL0 A0A1A8SLY6 A0A1J1IB09 A0A1A8M7E9
S4PN20 A0A1Y1MSL6 D6WV96 N6TVA1 A0A1W4WIT2 V9IIG1 A0A2A3EFN4 A0A088AW81 A0A232FG58 A0A067R6Z3 A0A026W1X4 A0A310SPY8 A0A0L7R5A7 E9IMQ6 A0A151XD95 A0A2J7PXJ2 A0A1B6JS33 A0A195D173 A0A0V0G4P5 A0A2P8YIF9 A0A069DTJ3 A0A1Z5L2Y9 A0A195FMY5 A0A158NH14 F4WST6 A0A2R5LGM7 A0A151J1Z5 A0A1B6GM89 A0A1B6KGF7 A0A131Y1S6 A0A1B6HSL5 A0A293MZW9 R4WD53 A0A0P4VS30 R4FLB3 E2AJE7 A0A0J7KUG9 W8CAD1 A0A1B6DC69 A0A0T6BEB5 A0A293LJM6 A0A0K8R5Z9 E2B2C5 A0A195B759 A0A1B0BLN9 A0A1A9XJC8 A0A0A1WL38 A0A3P9Q4X7 A0A3B3VP53 A0A2L2Y073 A0A0K8WJD2 A0A034WAT0 A0A0M3QTM9 A0A131XDK6 A0A3B3YZR5 A0A1E1XCS4 A0A087Y215 A0A2R7WWI9 A0A131YIZ0 A0A224YRS1 A0A0A9YZ97 A0A1A9URM4 A0A146M1E4 A0A1A9ZVR1 B4JP30 L7M000 B4Q8T1 A0A023GP71 A0A0J9R0V6 B4KJG7 A0A023FLI3 A0A0C9SE65 D3TNG6 A0A1A9WWC6 A0A3B3QUA3 X2J7P4 M4AU55 Q9VL02 A0A0K8TS53 A0A1L8E0D6 A0A1L8E056 B4HWK0 B4MUD1 B3MPQ5 B3N965 A0A1B0D1U9 B4G777 B4M8G5 B4NZ89 U5EKL0 A0A1A8SLY6 A0A1J1IB09 A0A1A8M7E9
Pubmed
19121390
26354079
22118469
23622113
28004739
18362917
+ More
19820115 23537049 28648823 24845553 24508170 30249741 21282665 29403074 26334808 28528879 21347285 21719571 23691247 27129103 20798317 24495485 25830018 26561354 25348373 28049606 28503490 26830274 28797301 25401762 26823975 17994087 25576852 22936249 26131772 20353571 29240929 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23542700 26109357 26109356 26369729 17550304
19820115 23537049 28648823 24845553 24508170 30249741 21282665 29403074 26334808 28528879 21347285 21719571 23691247 27129103 20798317 24495485 25830018 26561354 25348373 28049606 28503490 26830274 28797301 25401762 26823975 17994087 25576852 22936249 26131772 20353571 29240929 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23542700 26109357 26109356 26369729 17550304
EMBL
BABH01029542
BABH01029543
NWSH01002507
PCG68116.1
PCG68115.1
KQ459562
+ More
KPI99767.1 KQ461175 KPJ07809.1 AGBW02013278 OWR43668.1 GAIX01001112 JAA91448.1 GEZM01025369 JAV87490.1 KQ971363 EFA07759.1 APGK01050751 KB741176 KB632099 ENN73205.1 ERL88794.1 JR045916 AEY60084.1 KZ288256 PBC30530.1 NNAY01000310 OXU29307.1 KK852699 KDR18178.1 KK107487 QOIP01000006 EZA50067.1 RLU21481.1 KQ761783 OAD57013.1 KQ414654 KOC65941.1 GL764255 EFZ18112.1 KQ982294 KYQ58344.1 NEVH01020856 PNF21058.1 GECU01015492 GECU01005784 JAS92214.1 JAT01923.1 KQ976973 KYN06660.1 GECL01003107 JAP03017.1 PYGN01000572 PSN44036.1 GBGD01001892 JAC86997.1 GFJQ02007935 GFJQ02005589 JAV99034.1 JAW01381.1 KQ981387 KYN41970.1 ADTU01015432 GL888327 EGI62743.1 GGLE01004538 MBY08664.1 KQ980447 KYN16011.1 GECZ01016999 GECZ01006197 JAS52770.1 JAS63572.1 GEBQ01029454 JAT10523.1 GEFM01003364 JAP72432.1 GECU01030033 GECU01021061 GECU01007686 GECU01006353 GECU01003647 JAS77673.1 JAS86645.1 JAT00021.1 JAT01354.1 JAT04060.1 GFWV01020142 MAA44870.1 AK417398 BAN20613.1 GDKW01001210 JAI55385.1 ACPB03007378 GAHY01001493 JAA76017.1 GL439972 EFN66468.1 LBMM01003020 KMQ94062.1 GAMC01005863 GAMC01005861 JAC00695.1 GEDC01014019 JAS23279.1 LJIG01001277 KRT85663.1 GFWV01003964 MAA28694.1 GADI01007515 JAA66293.1 GL445130 EFN90162.1 KQ976574 KYM80092.1 JXJN01016521 GBXI01014553 JAC99738.1 IAAA01001283 IAAA01001284 LAA01538.1 GDHF01001339 JAI50975.1 GAKP01007742 GAKP01007741 JAC51210.1 CP012523 ALC39193.1 GEFH01004900 JAP63681.1 GFAC01002277 JAT96911.1 AYCK01006757 KK855798 PTY23906.1 GEDV01010511 JAP78046.1 GFPF01005528 MAA16674.1 GBHO01005262 GBRD01003356 JAG38342.1 JAG62465.1 GDHC01005837 JAQ12792.1 CH916372 EDV99455.1 GACK01007398 JAA57636.1 CM000361 EDX04496.1 GBBM01000150 JAC35268.1 CM002910 KMY89479.1 CH933807 EDW13547.1 GBBK01001746 JAC22736.1 GBZX01001392 JAG91348.1 CCAG010009466 EZ422968 ADD19244.1 AE014134 AHN54314.1 AY119493 AAF52903.1 AAM50147.1 AHN54315.1 GDAI01000419 JAI17184.1 GFDF01001945 JAV12139.1 GFDF01001946 JAV12138.1 CH480818 EDW52395.1 CH963857 EDW76057.1 CH902620 EDV32303.1 CH954177 EDV58500.1 AJVK01022376 CH479180 EDW28331.1 CH940654 EDW57491.1 CM000157 EDW88784.1 GANO01001770 JAB58101.1 HAEH01007558 HAEI01016454 SBS18923.1 CVRI01000042 CRK95625.1 HAEF01011776 SBR52935.1
KPI99767.1 KQ461175 KPJ07809.1 AGBW02013278 OWR43668.1 GAIX01001112 JAA91448.1 GEZM01025369 JAV87490.1 KQ971363 EFA07759.1 APGK01050751 KB741176 KB632099 ENN73205.1 ERL88794.1 JR045916 AEY60084.1 KZ288256 PBC30530.1 NNAY01000310 OXU29307.1 KK852699 KDR18178.1 KK107487 QOIP01000006 EZA50067.1 RLU21481.1 KQ761783 OAD57013.1 KQ414654 KOC65941.1 GL764255 EFZ18112.1 KQ982294 KYQ58344.1 NEVH01020856 PNF21058.1 GECU01015492 GECU01005784 JAS92214.1 JAT01923.1 KQ976973 KYN06660.1 GECL01003107 JAP03017.1 PYGN01000572 PSN44036.1 GBGD01001892 JAC86997.1 GFJQ02007935 GFJQ02005589 JAV99034.1 JAW01381.1 KQ981387 KYN41970.1 ADTU01015432 GL888327 EGI62743.1 GGLE01004538 MBY08664.1 KQ980447 KYN16011.1 GECZ01016999 GECZ01006197 JAS52770.1 JAS63572.1 GEBQ01029454 JAT10523.1 GEFM01003364 JAP72432.1 GECU01030033 GECU01021061 GECU01007686 GECU01006353 GECU01003647 JAS77673.1 JAS86645.1 JAT00021.1 JAT01354.1 JAT04060.1 GFWV01020142 MAA44870.1 AK417398 BAN20613.1 GDKW01001210 JAI55385.1 ACPB03007378 GAHY01001493 JAA76017.1 GL439972 EFN66468.1 LBMM01003020 KMQ94062.1 GAMC01005863 GAMC01005861 JAC00695.1 GEDC01014019 JAS23279.1 LJIG01001277 KRT85663.1 GFWV01003964 MAA28694.1 GADI01007515 JAA66293.1 GL445130 EFN90162.1 KQ976574 KYM80092.1 JXJN01016521 GBXI01014553 JAC99738.1 IAAA01001283 IAAA01001284 LAA01538.1 GDHF01001339 JAI50975.1 GAKP01007742 GAKP01007741 JAC51210.1 CP012523 ALC39193.1 GEFH01004900 JAP63681.1 GFAC01002277 JAT96911.1 AYCK01006757 KK855798 PTY23906.1 GEDV01010511 JAP78046.1 GFPF01005528 MAA16674.1 GBHO01005262 GBRD01003356 JAG38342.1 JAG62465.1 GDHC01005837 JAQ12792.1 CH916372 EDV99455.1 GACK01007398 JAA57636.1 CM000361 EDX04496.1 GBBM01000150 JAC35268.1 CM002910 KMY89479.1 CH933807 EDW13547.1 GBBK01001746 JAC22736.1 GBZX01001392 JAG91348.1 CCAG010009466 EZ422968 ADD19244.1 AE014134 AHN54314.1 AY119493 AAF52903.1 AAM50147.1 AHN54315.1 GDAI01000419 JAI17184.1 GFDF01001945 JAV12139.1 GFDF01001946 JAV12138.1 CH480818 EDW52395.1 CH963857 EDW76057.1 CH902620 EDV32303.1 CH954177 EDV58500.1 AJVK01022376 CH479180 EDW28331.1 CH940654 EDW57491.1 CM000157 EDW88784.1 GANO01001770 JAB58101.1 HAEH01007558 HAEI01016454 SBS18923.1 CVRI01000042 CRK95625.1 HAEF01011776 SBR52935.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000019118 UP000030742 UP000192223 UP000242457 UP000005203 UP000215335 UP000027135 UP000053097 UP000279307 UP000053825 UP000075809 UP000235965 UP000078542 UP000245037 UP000078541 UP000005205 UP000007755 UP000078492 UP000015103 UP000000311 UP000036403 UP000008237 UP000078540 UP000092460 UP000092443 UP000242638 UP000261500 UP000092553 UP000261480 UP000028760 UP000078200 UP000092445 UP000001070 UP000000304 UP000009192 UP000092444 UP000091820 UP000261540 UP000000803 UP000002852 UP000001292 UP000007798 UP000007801 UP000008711 UP000092462 UP000008744 UP000008792 UP000002282 UP000183832
UP000019118 UP000030742 UP000192223 UP000242457 UP000005203 UP000215335 UP000027135 UP000053097 UP000279307 UP000053825 UP000075809 UP000235965 UP000078542 UP000245037 UP000078541 UP000005205 UP000007755 UP000078492 UP000015103 UP000000311 UP000036403 UP000008237 UP000078540 UP000092460 UP000092443 UP000242638 UP000261500 UP000092553 UP000261480 UP000028760 UP000078200 UP000092445 UP000001070 UP000000304 UP000009192 UP000092444 UP000091820 UP000261540 UP000000803 UP000002852 UP000001292 UP000007798 UP000007801 UP000008711 UP000092462 UP000008744 UP000008792 UP000002282 UP000183832
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JHV1
A0A2A4J8M5
A0A2A4J7Q1
A0A194Q2H7
A0A194QR36
A0A212EQA7
+ More
S4PN20 A0A1Y1MSL6 D6WV96 N6TVA1 A0A1W4WIT2 V9IIG1 A0A2A3EFN4 A0A088AW81 A0A232FG58 A0A067R6Z3 A0A026W1X4 A0A310SPY8 A0A0L7R5A7 E9IMQ6 A0A151XD95 A0A2J7PXJ2 A0A1B6JS33 A0A195D173 A0A0V0G4P5 A0A2P8YIF9 A0A069DTJ3 A0A1Z5L2Y9 A0A195FMY5 A0A158NH14 F4WST6 A0A2R5LGM7 A0A151J1Z5 A0A1B6GM89 A0A1B6KGF7 A0A131Y1S6 A0A1B6HSL5 A0A293MZW9 R4WD53 A0A0P4VS30 R4FLB3 E2AJE7 A0A0J7KUG9 W8CAD1 A0A1B6DC69 A0A0T6BEB5 A0A293LJM6 A0A0K8R5Z9 E2B2C5 A0A195B759 A0A1B0BLN9 A0A1A9XJC8 A0A0A1WL38 A0A3P9Q4X7 A0A3B3VP53 A0A2L2Y073 A0A0K8WJD2 A0A034WAT0 A0A0M3QTM9 A0A131XDK6 A0A3B3YZR5 A0A1E1XCS4 A0A087Y215 A0A2R7WWI9 A0A131YIZ0 A0A224YRS1 A0A0A9YZ97 A0A1A9URM4 A0A146M1E4 A0A1A9ZVR1 B4JP30 L7M000 B4Q8T1 A0A023GP71 A0A0J9R0V6 B4KJG7 A0A023FLI3 A0A0C9SE65 D3TNG6 A0A1A9WWC6 A0A3B3QUA3 X2J7P4 M4AU55 Q9VL02 A0A0K8TS53 A0A1L8E0D6 A0A1L8E056 B4HWK0 B4MUD1 B3MPQ5 B3N965 A0A1B0D1U9 B4G777 B4M8G5 B4NZ89 U5EKL0 A0A1A8SLY6 A0A1J1IB09 A0A1A8M7E9
S4PN20 A0A1Y1MSL6 D6WV96 N6TVA1 A0A1W4WIT2 V9IIG1 A0A2A3EFN4 A0A088AW81 A0A232FG58 A0A067R6Z3 A0A026W1X4 A0A310SPY8 A0A0L7R5A7 E9IMQ6 A0A151XD95 A0A2J7PXJ2 A0A1B6JS33 A0A195D173 A0A0V0G4P5 A0A2P8YIF9 A0A069DTJ3 A0A1Z5L2Y9 A0A195FMY5 A0A158NH14 F4WST6 A0A2R5LGM7 A0A151J1Z5 A0A1B6GM89 A0A1B6KGF7 A0A131Y1S6 A0A1B6HSL5 A0A293MZW9 R4WD53 A0A0P4VS30 R4FLB3 E2AJE7 A0A0J7KUG9 W8CAD1 A0A1B6DC69 A0A0T6BEB5 A0A293LJM6 A0A0K8R5Z9 E2B2C5 A0A195B759 A0A1B0BLN9 A0A1A9XJC8 A0A0A1WL38 A0A3P9Q4X7 A0A3B3VP53 A0A2L2Y073 A0A0K8WJD2 A0A034WAT0 A0A0M3QTM9 A0A131XDK6 A0A3B3YZR5 A0A1E1XCS4 A0A087Y215 A0A2R7WWI9 A0A131YIZ0 A0A224YRS1 A0A0A9YZ97 A0A1A9URM4 A0A146M1E4 A0A1A9ZVR1 B4JP30 L7M000 B4Q8T1 A0A023GP71 A0A0J9R0V6 B4KJG7 A0A023FLI3 A0A0C9SE65 D3TNG6 A0A1A9WWC6 A0A3B3QUA3 X2J7P4 M4AU55 Q9VL02 A0A0K8TS53 A0A1L8E0D6 A0A1L8E056 B4HWK0 B4MUD1 B3MPQ5 B3N965 A0A1B0D1U9 B4G777 B4M8G5 B4NZ89 U5EKL0 A0A1A8SLY6 A0A1J1IB09 A0A1A8M7E9
PDB
6G2V
E-value=6.57813e-09,
Score=138
Ontologies
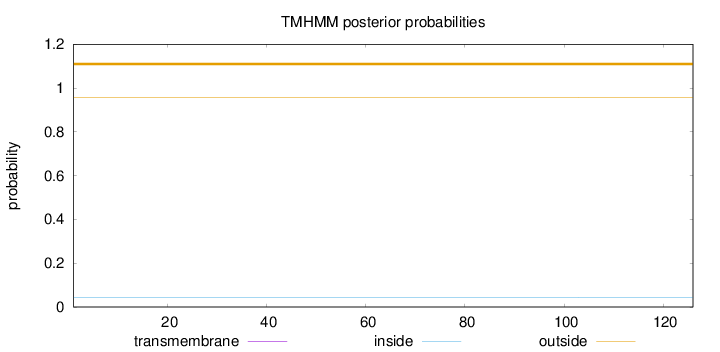
Topology
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04430
outside
1 - 126
Population Genetic Test Statistics
Pi
10.131918
Theta
17.750308
Tajima's D
-1.305635
CLR
331.191874
CSRT
0.0888955552222389
Interpretation
Uncertain
